Family: Caulimoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are either isometric or bacilliform depending on the genus (Figure 1). There is no envelope.
Physicochemical and physical properties
Virions have buoyant densities in CsCl of 1.37 g cm−3 (genera with isometric virions) or in Cs2SO4 of 1.31 g cm−3 (genera with bacilliform virions). S20,w is in the range of 200S to 220S. Particles are very stable between pH4 and pH9 and in high salt concentrations.
Nucleic acid
Virions contain a single molecule of non-covalently closed circular dsDNA of 7.2–9.2 kbp. Each strand of the genome has discontinuities at specific places: the minus-strand has one discontinuity and the plus-strand has between one and three discontinuities.
Proteins
Genomes contain between one and eight ORFs, depending on the genus. The virus-encoded proteins common to all genera are a movement protein, a coat protein, a multipurpose virion-associated protein, an aspartic protease and a reverse transcriptase (RT) with associated RNAse H1 activity.
Lipids
None reported.
Carbohydrates
The coat protein of cauliflower mosaic virus (CaMV) is glycosylated. No carbohydrates have been reported for the virions of other species.
Genome organization and replication
By convention, numbering of the genome starts at the 5’ end of the minus-strand primer-binding site. One strand of DNA contains the coding sequence (plus-strand). The genome organization is dependent upon the genus (Figure 2) and is one of the main characteristics that distinguish the genera from each other.
Following entry into the cell, the virion is targeted to the nucleus by a nuclear localization signal (NLS) that is located in the N-terminus of the coat protein and is exposed on the surface of the virion. It is then thought that the virion docks at a nuclear pore, virion disassembly occurs and the DNA is imported into the nucleus utilizing the importin α pathway or perhaps other pathways in the case of rice tungro bacilliform virus (RTBV). The discontinuities in the genome are then sealed to give supercoiled DNA, which associates with histone proteins to form minichromosomes in the nucleus. These are transcribed asymmetrically by host DNA-dependent RNA polymerases to give a greater-than-genome length transcript (35S or 34S RNA) that has a terminal redundancy of about 35 to 270 nt, dependent upon the species. This transcript serves as a template (the pregenomic RNA) for reverse transcription to give the minus-strand DNA and as a polycistronic mRNA for expression of at least some of the ORFs.
The 5’ leader sequence of the pregenomic RNA of nearly all members of the Caulimoviridae is long and folds into a large and stable hairpin structure. Immediately preceding this hairpin structure is a small ORF (sORFA). Initiation of translation of ORF1 occurs by a ribosome shunt mechanism, whereby translation begins at sORFA but then the ribosome is shunted across the secondary structure to a landing site near the beginning of ORF1. Viruses in the genera Caulimovirus and Soymovirus produce a specific monocistronic mRNA (19S RNA) for ORF6; no sgRNAs have been reported for genera Petuvirus, Soymovirus, Cavemovirus and Badnavirus. ORF4 of RTBV is expressed from an RNA spliced from the pregenomic RNA.
The replication cycle, in contrast to that of retroviruses, is episomal and does not involve an integration phase. Minus-strand DNA synthesis is primed by host cytosolic tRNAmet and synthesis of both strands is performed by the viral RT and RNAse H1. The site-specific discontinuities are at the priming sites for both minus- and plus-strand DNA synthesis and are made by the oncoming strand displacing the existing strand for a short distance and not ligating to form a closed circle.
Antigenic properties
Virions range from moderate to efficient immunogens. There is pronounced antigenic variability within species in the genus Badnavirus. There are some serological cross-reactions between members of different genera.
Biological properties
The host ranges of most species are narrow. Those in the genera Petuvirus, Soymovirus and Cavemovirus are restricted to dicotyledonous plants; tungroviruses infect monocotyledonous plants and badnaviruses infect either dicotyledonous or monocotyledonous plants. Many members of the family are spread by vegetative propagation.
The geographic range of many species is wide; most species in the genera Tungrovirus and Badnavirus are primarily tropical or subtropical with some temperate and sub-Antarctic species whereas most of the species in the genera Petuvirus, Caulimovirus, Soymovirus and Cavemovirus are found in temperate regions.
The symptoms caused by these viruses are variable and dependent on the virus species, host and climatic conditions. Mosaic or vein clearing symptoms predominate amongst members of the genera Petuvirus, Caulimovirus, Soymovirus and Cavemovirus, whereas interveinal chlorotic mottling and streaking is the most frequent symptom of those in the genera Tungrovirus and Badnavirus.
Most viruses in the family infect most cell types of their hosts although some in the genera Tungrovirus and Badnavirus are restricted to the vascular system. Virions occur in the cytoplasm and those of species in the genera Petuvirus, Caulimovirus, Soymovirus and Cavemovirus are associated with virus-encoded proteinaceous inclusion bodies.
Members of the genera Caulimovirus, Petuvirus, Cavemovirus and Badnavirus may have both endogenous (viral DNA integrated in the host nuclear genome) and exogenous forms. Integration is not an obligatory step in the replication cycles of these viruses but rather the DNA has become captured in the host nuclear genome by non-homologous end-joining (also known as illegitimate recombination). Replication-competent endogenous caulimovirid sequences occur in Musa balbisiana, Petunia hybrida and Nicotiana edwardsonii. Replication-defective endogenous caulimovirid sequences are found in many other dicotyledonous and monocotyledonous plant species.
Genus Caulimovirus
Type species Cauliflower mosaic virus
Distinguishing features
Members of this genus have particles and cytoplasmic inclusions similar to those of members of the genera Soymovirus, Petuvirus and Cavemovirus but differ from them in genome organization and phylogenetic placement using polymerase gene sequences. Caulimoviruses have six open reading frames and can be distinguished from their closest relatives, the soymoviruses, by the presence of only two ORFs between ORF1 (movement protein) and ORF4 (coat protein) instead of three. The position of the minus-strand primer-binding site also differs between the two genera, as does the presence of an intergenic region between ORF5 and 6.
Virion properties
Morphology
Isometric virions are 52 nm in diameter with an icosahedral T7 symmetry. The virion capsid consists of three concentric shells, built from 420 P4 proteins assembled as 60 hexavalent and 12 pentavalent capsomers. The inner cavity has a diameter of about 25 nm. P3 proteins are incorporated in the virion as a triskelion structure that cements three hexavalent or pentavalent capsomers together. The N-terminus of the P3 protein, containing two coiled-coil motifs with opposite handedness, is exposed at the surface of the virion. The C-terminus of the P3 protein, which has a nucleic acid-binding motif, is embedded inside the pores surrounding the capsomers and traverses the P4 protein layers to reach the genomic DNA.
Physicochemical and physical properties
Virions have a buoyant density of 1.35–1.38 g cm−3 in CsCl. Sedimentation coefficients are between 215 and 245 S.
Nucleic acid
Virions contain a single molecule of non-covalently closed circular dsDNA of about 7.8–8.2 kbp. The minus-strand DNA has a single discontinuity and the plus-strand DNA, two or three discontinuities.
Genome organization and replication
The genome contains six ORFs and large and small intergenic regions. The large intergenic region, containing the pregenomic RNA (35S) promoter, the RNA polyadenylation signal and the minus-strand primer-binding site, is located between ORF6 and ORF1. The small intergenic region, containing the 19S promoter, is located between ORF5 and 6. Protein P1 is the movement protein, P2 is the aphid transmission factor, P3 is the virion-associated protein, P4 is the coat protein precursor, P5 is the polymerase polyprotein (aspartic protease, RT and RNaseH1 enzymatic activities) and P6 is the translation transactivator/viral silencing suppressor. Protein P4 of CaMV is processed by the viral aspartic protease at both the N- and C-termini to yield three major polypeptides of ca. 44, 39 and 37 kDa. The N-terminus of the 44 kDa polypeptide is at alanine 77 and the C-terminus between positions 435 and 440.
Electron dense inclusion bodies (the viroplasm), which are composed primarily of P6, occur in the cytoplasm and are the site of viral DNA and protein synthesis, morphogenesis and storage of virus particles. Single electron lucent transmission bodies (TBs) are also found in the cytoplasm of each infected cell and are composed of P2 and P3 proteins. These TBs are ingested by the aphid vector and in an interaction involving the N-terminus of the P2 protein, bind to the tip of the aphid stylet. The P3 protein is then released and the bound P2 protein then binds P3-decorated virions during subsequent intracellular probes by the aphid.
Two major capped and polyadenylated transcripts (35S and 19S RNAs) are produced. The 35S RNA serves as the template for reverse transcription and as a polycistronic mRNA for proteins P1 to P5. P6 is transcribed from the 19S RNA and enables re-initiation of translation of downstream ORFs in a polycistronic mRNA after stop codons are passed. Several spliced versions of the 35S RNA are generated, some containing ORF3 and downstream sequences and others, ORF1 and 2 fused in-frame. This splicing is thought to prevent excessive expression of P2, which is inhibitory to virus replication. As much as 70% of the total viral RNA population is spliced.
Antigenic properties
The viruses are moderately to strongly immunogenic. Serological cross-reactivity between the isolates of different species is moderate to strong.
Biological properties
The viruses are transmitted in a semi-persistent manner by aphids; LLDV has no known vector. Transmission requires a virus-encoded protein (aphid transmission factor) that is encoded by ORF2. All except SVBV and LLDV are mechanically transmissible. Seed transmission is not recorded.
Natural host ranges are narrow (restricted to a single plant family) although some species have been experimentally transmitted to hosts in two to four plant families. Hosts are limited to dicotyledonous plants.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Differences in host ranges
- Differences in polymerase (RT+RNAse H) nt sequences of more than 20%
- Differences in gene product sequences.
List of species in the genus Caulimovirus
| Carnation etched ring virus |
|
|
| Carnation etched ring virus-UK | [X04658=NC_003498] | (CERV-UK) |
| Cauliflower mosaic virus |
|
|
| Cauliflower mosaic virus-Cabb-S | [V00141=NC_001497] | (CaMV-CabbS) |
| Dahlia mosaic virus |
|
|
| Dahlia mosaic virus-USA | [AY309479*] | (DMV-US) |
| Figwort mosaic virus |
|
|
| Figwort mosaic virus-USA | [X06166=NC_003554] | (FMV-US) |
| Horseradish latent virus |
|
|
| Horseradish latent virus-Denmark | [AY534728-33*] | (HRLV-DK) |
| Mirabilis mosaic virus |
|
|
| Mirabilis mosaic virus-Illinois | [AF454635=NC_004036] | (MiMV_IL) |
| Strawberry vein banding virus |
|
|
| Strawberry vein banding virus-Czech Republic | [X97304=NC_001725] | (SVBV-CR) |
| Thistle mottle virus |
|
|
| Thistle mottle virus-UK |
| (ThMoV-UK) |
Species names are in italic script; names of isolates and synonyms are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Caulimovirus but have not been approved as species
| Aquilegia necrotic mosaic virus |
| (ANMV) |
| Dahlia common mosaic virus |
| (DCMV) |
| Eupatorium vein clearing virus | [EU569831=NC_010738] | (EVCV) |
| Lamium leaf distortion virus | [EU554423=NC_010737] | (LLDV) |
| Plantago virus 4 |
| (PlV-4) |
| Rudbeckia flower distortion virus | [FJ493469=NC_011920] | (RuFDV) |
| Sonchus mottle virus |
| (SMoV) |
Genus Petuvirus
Type species Petunia vein clearing virus
Distinguishing features
Petunia vein clearing virus (PVCV), the sole member of the genus, is distinguishable from all other members of the Caulimoviridae by its simple genome organization (one ORF) and phylogenetic placement using polymerase gene sequences.
Virion properties
Morphology
Virions are isometric in shape.
Physicochemical and physical properties
No information available.
Nucleic acid
Virions contain a single molecule of non-covalently closed circular dsDNA of about 7.2 kbp.
Genome organization and replication
The genome contains a single ORF encoding a large polyprotein and an intergenic region containing the pregenomic RNA promoter, a polyadenylation signal and the minus-strand primer-binding site. Protein domains characteristic of the movement protein, coat protein, aspartic protease, reverse transcriptase and RNaseH1 are present in the ORF1 polyprotein in that order. There is also a putative virion-associated protein homolog between the movement protein and coat protein domains of the polyprotein.
The 5’ leader sequence of the pregenomic RNA folds into a stable hairpin structure preceded by a small ORF. Thus a ribosome shunt mechanism of initiation of translation is assumed to be utilized. The polyprotein is processed by the aspartic protease to generate mature proteins. How the timing and level of expression of the different proteins is coordinated during the replication cycle is unknown. Infected cells contain electron dense inclusion bodies similar to those produced by infections of members of the genus Caulimovirus.
Antigenic properties
No information available.
Biological properties
Infection in Petunia hybrida is due to activation of replication-competent endogenous PVCV (ePVCV) sequences in the nuclear genome of the plant. Wounding (e.g. plant pruning) induces infection. The virus is vertically transmitted to all progeny as part of the host’s chromosomes and is also graft-transmissible to Petunia parodii and Nicotiana glutinosa. PVCV has no known insect vectors and it is not mechanically transmissible.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Petuvirus
| Petunia vein clearing virus |
|
|
| Petunia vein clearing virus-USA | [U95208=NC_001839] | (PVCV-US) |
Species name is in italic script; name of isolate is in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Petuvirus but have not been approved as species
None reported.
Genus Soymovirus
Type species Soybean chlorotic mottle virus
Distinguishing features
Members of this genus have particles and cytoplasmic inclusions similar to those of species in the genera Caulimovirus, Petuvirus and Cavemovirus but differ from them in genome organization and phylogenetic placement using polymerase gene sequences. Soymoviruses have seven or eight ORFs and can be distinguished from their closest relatives in the genus Caulimovirus by the presence of three ORFs (ORF1b, ORF2 and ORF3) between the movement protein and coat protein-coding ORFs. The minus-strand primer-binding site is also located in ORF1b or in a small intergenic region immediately downstream of this ORF. Furthermore, there is no intergenic region between ORF5 and ORF6 as there is with members of the genus Caulimovirus.
Virion properties
Morphology
Virions are isometric and about 50 nm in diameter.
Physicochemical and physical properties
When virion preparations of blueberry red ringspot virus-USA (BRRSV) are centrifuged in sucrose or CsCl gradients, two components are observed with buoyant densities in CsCl of 1.30 and 1.40 g cm−3. Virions from the two fractions have no morphological differences. A0.1%, 1cm, 260nm about 7.0.
Nucleic acid
Virions contain a single molecule of non-covalently closed circular dsDNA of about 8.1–8.3 kbp. The minus-strand DNA has a single discontinuity and the plus-strand DNA, two discontinuities.
Genome organization and replication
The genome contains seven or eight ORFs (ORFs 1a, 1b, 2–7). There is one large intergenic region between ORF6 and 7, in which is located the pregenomic RNA promoter and the polyadenylation signal. Additionally, for BRRSV and Cestrum yellow leaf curling virus (CmYLCV) there is a small intergenic region downstream of ORF1b. The location of the minus-strand primer-binding site is either within ORF1b (soybean chlorotic mottle virus [SbCMV] and peanut chlorotic streak virus] or in the small intergenic region downstream of ORF1b [BRRSV and CmYLCV]). ORF1b and ORF7 are nonessential for replication and systemic infection and whether they are expressed is unknown. Furthermore, ORF7 is absent in CmYLCV. Protein P1a is the movement protein, the function of P2 is unknown, P3 is the virion-associated protein, P4 is the coat protein precursor, P5 is the polymerase polyprotein (aspartic protease, RT and RNaseH enzymatic activities) and P6 is the translation transactivator.
Two major RNA transcripts, the first representing the pregenomic RNA (c. 8.2 kbp) and the second, a monocistronic RNA (1.8 kbp) containing ORF6, have been observed for SbCMV. No putative promoter sequences could be identified upstream of the 5’-end of the smaller RNA species. The pregenomic RNA serves as a polycistronic mRNA and protein P6 enables expression of the downstream ORFs. Electron dense inclusion bodies (the viroplasm) are visible in infected cells and are likely to fulfil a similar role to those of members of the genus Caulimovirus. The 5’ leader sequence of the pregenomic RNA of SbCMV does not fold into a strong secondary structure as for other members of the family Caulimoviridae, and whether a ribosome shunt mechanism of initiation of translation of ORF1 is utilized requires further investigation.
Antigenic properties
Virions are moderately immunogenic if fixed in 0.5% (v/v) formaldehyde. Serological relationships between species in this genus are unknown but no cross-reactivity with species in the genus Caulimovirus has been observed.
Biological properties
Host ranges are narrow (one to two plant families). All but BRRSV are mechanically transmissible. Spread of the viruses in the field is observed although the vectors are unknown. BRRSV is spread by clonal propagation of its host.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Differences in host ranges
- Differences in polymerase (RT+RNAse H) nt sequences of more than 20%
- Differences in gene product sequences.
List of species in the genus Soymovirus
| Blueberry red ringspot virus |
|
|
| Blueberry red ringspot virus-USA | [AF404509=NC_003138] | (BRRSV-US) |
| Peanut chlorotic streak virus |
|
|
| Peanut chlorotic streak virus-K1 | [U13988=NC_001634] | (PCSV-K1) |
| Soybean chlorotic mottle virus |
|
|
| Soybean chlorotic mottle virus-Japan | [X15828=NC_001739] | (SbCMV-JA) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Soymovirus but have not been approved as species
| Cestrum yellow leaf curling virus | [AF364175=NC004324] | (CmYLCV) |
Genus Cavemovirus
Type species Cassava vein mosaic virus
Distinguishing features
The members of this genus produce particles and cytoplasmic inclusions similar to those of species in the genera Caulimovirus, Petuvirus and Soymovirus but differ from them in genome organization and phylogenetic placement using polymerase gene sequences. The order of the movement protein and coat protein is also reversed to that of all other genera in the Caulimoviridae.
There are subtle differences in the genome organization of cassava vein mosaic virus (CsVMV) and tobacco vein clearing virus (TVCV), and the genetic distance between the two in the polymerase polyprotein gene is of a similar magnitude to different genera in the Caulimoviridae. A proposal to split these two species into different genera is currently under consideration.
Virion properties
Morphology
Virions are isometric and about 50 nm in diameter.
Physicochemical and physical properties
TVCV virions have a buoyant density of 1.35 g cm−3 in Cs2SO4. The sedimentation coefficient, S20,w, of CsVMV is estimated to be about 246S.
Nucleic acid
Virions contain a single molecule of non-covalently closed circular dsDNA of about 7.7–8.2 kbp.
Genome organization and replication
Both CsVMV and TVCV have four ORFs and a large intergenic region, in which is located the pregenomic RNA promoter, the RNA polyadenylation signal and the minus-strand primer-binding site. ORF1 of CsVMV encodes a polyprotein with coat protein and movement protein domains in that order. In contrast, the coat protein and movement protein of TVCV are encoded by separate ORFs (ORFs 1 and 2). Furthermore, the CsVMV ORF1 protein has a 124-aa N-terminal extension of unknown function relative to the TVCV ORF1 protein. The putative ORF2 protein of CsVMV is small (8.8 kDa), has no known function and there is no homolog in the TVCV genome. For both viruses, the proteins encoded by ORF3 and 4 are the polymerase polyprotein (aspartic protease, RT and RNase H1 enzymatic activities) and the virion-associated protein/translation transactivator, respectively.
The 5’ leader sequence of the pregenomic RNA folds into a stable hairpin structure preceded by a small ORF and thus a ribosome shunt mechanism of initiation of translation of ORF1 is assumed to be utilized. The presence of a translation transactivator homolog in the genome would suggest a similar mechanism of expression of downstream ORFs in a multicistronic mRNA as utilized by members of the genus Caulimovirus.
Antigenic properties
No information available.
Biological properties
There is no conclusive evidence of vector or seed transmission of CsVMV. The only known host of this virus is cassava (Manihot esculenta). The virus is transmitted by vegetative propagation.
Attempts at mechanical, graft or aphid transmission of TVCV have been unsuccessful. Infection in the interspecific allohexaploid Nicotiana edwardsonii (N. clevelandii×N. glutinosa) is believed to originate from activation of replication-competent endogenous TVCV (eTVCV) sequences in the nuclear genome of the plant. The virus is vertically transmitted to all progeny as part of the host’s chromosomes. TVCV infections have only ever been observed in Nicotiana edwardsonii although eTVCV sequences are present in Nicotiana sylvestris, Nicotiana tabacum, Nicotiana tomentosiformis, Solanum habrochaites and Solanum lycopersicum and related sequences have been detected in several other plant species in the Solanaceae.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Differences in host ranges
- Differences in polymerase (RT+RNAse H) nt sequences of more than 20%
- Differences in gene product sequences.
List of species in the genus Cavemovirus
| Cassava vein mosaic virus |
|
|
| Cassava vein mosaic virus-Brazil | [U59751=NC_001648] | (CsVMV-BR) |
| Tobacco vein clearing virus |
|
|
| Tobacco vein clearing virus-USA | [AF190123=NC_003378] | (TVCV-US) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Cavemovirus but have not been approved as species
None reported.
Genus Badnavirus
Type species Commelina yellow mottle virus
Distinguishing features
The genera Badnavirus and Tungrovirus are unique among the family Caulimoviridae in having bacilliform-shaped virions. Members of the genus Badnavirus can be distinguished from rice tungro bacilliform virus (RTBV), the sole member of the genus Tungrovirus, by genome organization, the lack of any RNA splicing during replication, the lack of dependency on a helper virus for vector transmission, and phylogenetic placement using polymerase gene sequences.
Virion properties
Morphology
Virions are bacilliform with parallel sides and rounded ends (Figure 1). Virions are uniformly 30 nm in width. The modal particle length is 130 nm, but particles ranging in length from 60 to 900 nm are commonly observed. No projections or other capsid surface features have been observed by electron microscopy. The tubular portion of the virion has a structure based on an icosahedron cut across its 3-fold axis, with a structural repeat of 10 nm and nine rings of hexamer subunits per 130 nm length.
Physicochemical and physical properties
Purified virions have an A260/280nm ratio of 1.26 (uncorrected for light scattering).
Nucleic acid
Virions contain a single molecule of non-covalently closed circular dsDNA of about 7.2–9.2 kbp. Each strand of the genome has a single discontinuity.
Genome organization and replication
The genome contains three ORFs. The function of protein P1 is unknown, P2 is the virion-associated protein, P3 is a polyprotein with movement protein, coat protein, aspartic protease and RT/RNase H1 domains in that order.
A single, greater-than-genome length, terminally redundant pregenomic RNA is transcribed. No subgenomic RNAs have been observed. The pregenomic RNA serves as a polycistronic mRNA for translation of the three ORFs. By analogy to RTBV, translation of ORF1 is initiated by ribosome shunting and translation of ORF2 and 3 by leaky scanning. Consistent with a leaky scanning model of translation, the start codon of ORF1 and ORF2 are in unfavourable translation contexts and there is paucity of internal AUG codons in both ORFs. In banana streak MY virus and banana streak VN virus, ORF1 begins with a nonconventional start codon (CUG). Furthermore, ORF3 is in −1 translational frame relative to ORF2, which in turn is in a −1 translational frame relative to ORF1. ORFs 1 and 2 and 2 and 3 also have overlapping start and stop codons (ATGA).
Sites where the ORF3 polyprotein is cleaved by the viral aspartic protease have not been determined.
Antigenic properties
Serological relationships between species members are weak to absent.
Biological properties
Transmission is in a semi-persistent manner by mealybugs and for some members, by aphids or lacebugs. The virus does not multiply in its mealybug vector and there is no transovarial transmission. All motile life stages of vectors can acquire and transmit the virus. There is little information on the possible transmission of badnaviruses by other vector types. Seed transmission at a rate of 30–63% has been recorded for Kalanchoë top-spotting virus (KTSV). KTSV, Cacao swollen shoot virus and an unidentified badnavirus from sugarcane have been mechanically transmitted but attempts with other species have been unsuccessful, which may relate to the presence of inhibitory substances in the plant sap or low virus titres. Some species in woody hosts have been transmitted by dodder and grafting. Biological host ranges are narrow and restricted to one or two plant families.
Replication-competent endogenous badnaviral sequences (banana streak OL, banana streak GF and probably banana streak MY viruses) are present in Musa balbisiana. These endogenous sequences are responsible for causing infection in M. acuminata × M. balbisiana hybrids, especially following plant propagation by tissue culture. Many other plant species contain endogenous badnaviral sequences but most are probably replication-defective.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Differences in host ranges
- Differences in polymerase (RT+RNAse H) nt sequences of more than 20%
- Differences in gene product sequences
- Differences in vector specificities.
List of species in the genus Badnavirus
| Aglaonema bacilliform virus |
|
|
| Aglaonema bacilliform virus |
| (ABV) |
| Banana streak GF virus |
|
|
| Banana streak GF virus-Ecuador | [AY493509=NC_007002] | (BSGFV-EC) |
| Banana streak Mysore virus |
|
|
| Banana streak MY virus-Australia | [AY805074=NC_006955] | (BSMYV-AUS) |
| Banana streak OL virus |
|
|
| Banana streak OL virus-Nigeria | [AJ002234=NC_003381] | (BSOLV-NI) |
| Cacao swollen shoot virus |
|
|
| Cacao swollen shoot virus-Agou1 | [L14546=NC_001574] | (CSSV-AGOU1) |
| Canna yellow mottle virus |
|
|
| Canna yellow mottle virus-Italy1 | [EF156357*] | (CaYMV-IT1) |
| Citrus mosaic virus |
|
|
| (Citrus yellow mosaic virus) |
|
|
| Citrus mosaic virus-India | [AF347695=NC_003382] | (CiMV-IN) |
| Commelina yellow mottle virus |
|
|
| Commelina yellow mottle virus-Guadeloupe | [X52938=NC_001343] | (ComYMV-GU) |
| Dioscorea bacilliform virus |
|
|
| Dioscorea bacilliform Al virus-L85 | [X94576*] | (DBALV-L85) |
| Gooseberry vein banding associated virus |
|
|
| Gooseberry vein banding associated virus GB1 | [HQ852248] | (GVBAV-SCO) |
| Kalanchoë top-spotting virus |
|
|
| Kalanchoë top-spotting virus-USA | [AY180137=NC_004540] | (KTSV-US) |
| Piper yellow mottle virus |
|
|
| Piper yellow mottle virus-Sri Lanka | [AJ626981*] | (PYMoV-SRL) |
| Rubus yellow net virus |
|
|
| Rubus yellow net virus-UK | [AF468454*] | (RYNV) |
| Schefflera ringspot virus |
|
|
| Schefflera ringspot virus-USA |
| (SRV-US) |
| Spiraea yellow leaf spot virus |
|
|
| Spiraea yellow leaf spot virus-USA | [AF299074*] | (SYLSV_US) |
| Sugarcane bacilliform IM virus |
|
|
| Sugarcane bacilliform IM virus-Queensland | [AJ277091=NC_003031] | (SCBIMV-QLD) |
| Sugarcane bacilliform Mor virus |
|
|
| Sugarcane bacilliform Mor virus-Morocco | [M89923=NC_008017] | (SCBMV-MOR) |
| Taro bacilliform virus |
|
|
| Taro bacilliform virus-Papua New Guinea | [AF357836=NC_004450] | (TaBV-PNG) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Badnavirus but have not been approved as species
| Aucuba bacilliform virus |
| (AuBV) |
| Banana streak VN virus | [AY750115=NC_007003] | (BSVNV) |
| Bougainvillea chlorotic vein banding virus | [EU034539=NC_011592] | (BCVBV) |
| Dioscorea bacilliform SN virus | [DQ822073=NC_009010] | (DBSNV) |
| Dracaena mottle virus | [DQ473478=NC_008034†] | (DrMV) |
| Mimosa bacilliform virus |
| (MBV) |
| Pelargonium vein banding virus | [GQ428155=NC_13262] | (PlVBV) |
| Pineapple bacilliform CO virus | [EU377666*] | (PBCOV) |
| Pineapple bacilliform ER virus | [EU377673*] | (PBERV) |
| Stilbocarpa mosaic bacilliform virus | [AF478961*] | (SMBV) |
| Sweetpotato badnavirus A | [FJ560943] |
|
| Sweetpotato badnavirus B | [FJ560944=NC_012728] |
|
| Yucca bacilliform virus | [AF468688*†] | (YBV) |
* Sequences do not comprise the complete genome.
† Sequence amplified by PCR from total plant DNA extract; it has not been proved that the sequence derives from an exogenous viral genome.
Genus Tungrovirus
Type species Rice tungro bacilliform virus
Distinguishing features
RTBV is the sole member of the genus Tungrovirus. Along with members of the genus Badnavirus, RTBV is readily distinguished from other genera in the Caulimoviridae by its bacilliform-shaped virions. Although the genome organization of RTBV is very similar to that of the genus Badnavirus, it differs by the presence of a fourth ORF (ORF4). This fourth ORF is expressed from a monocistronic RNA that is produced by splicing of the pregenomic RNA. RTBV can also be differentiated from members of the genus Badnavirus by phylogenetic placement using polymerase gene sequences.
Virion properties
Morphology
The virus particles are bacilliform of diameter 30 nm and predominant length 130 nm; longer particles, up to 300 nm, are found in some isolates. The particle structure is based on a T=3 icosahedron cut across its three-fold axis, the tubular portion being made up of rings of hexamer subunits with a repeat distance of about 10 nm.
Physicochemical and physical properties
The particles sediment with an S20,w of about 200S and have a buoyant density in CsCl of about 1.36 g ml−1.
Nucleic acid
Virions contain a single molecule of non-covalently closed circular dsDNA of about 8.0 kbp. Each strand of the genome has a single discontinuity.
Genome organization and replication
The genome contains four ORFs, a large intergenic region between ORF4 and 1 and a small intergenic region between ORF3 and 4. The pregenomic RNA promoter, polyadenylation signal and minus-strand primer-binding site are all located in the large intergenic region. The function of protein P1 is unknown, P2 is the virion-associated protein, P3 is a polyprotein with movement protein, coat protein, aspartic protease and RT/RNase H1 domains in that order and the function of protein P4 is unknown.
A single, greater-than-genome length, terminally redundant pregenomic RNA is transcribed. The pregenomic RNA serves as a polycistronic mRNA for translation of ORFs 1-3. Translation of ORF1 is initiated by ribosome shunting and translation of ORF2 and 3 by leaky scanning. Consistent with a leaky scanning model of translation, the start codon of ORF1 is non-conventional (AUU), the start codon of ORF2 is in an unfavourable translation context and there is an absence of internal AUG codons in both ORFs. Furthermore, ORF3 is in −1 translational frame relative to ORF2, which in turn is in a −1 translational frame relative to ORF1. ORFs 1 and 2 and 2 and 3 also have overlapping start and stop codons (ATGA).
ORF4 is expressed from a monocistronic RNA that is produced by splicing of the pregenomic RNA. A 6300 nt intron is excised in a way that fuses the first small ORF (sORFA) in the leader sequence of the pregenomic RNA in frame to ORF4 at a position that is 22 codons upstream of the start codon of this ORF.
Protein precursors in the ORF3 polyprotein are processed through the action of the viral aspartic protease. The mature coat protein has been mapped to aa 477–791 (GenBank accession NP_056762), giving rise to a protein of 37.3 kDa. The aspartic protease has been mapped to aa 965–1085, giving rise to a protein of 13.8 kDa.
Antigenic properties
RTBV is moderately immunogenic.
Biological properties
RTBV is transmitted by leafhoppers in the genera Nephotettix and Recilia. Although able to replicate independently in its host, RTBV is only able to be transmitted when the leafhopper has acquired rice tungro spherical virus (genus Waikavirus) simultaneously or previously, suggesting that RTSV may contribute a helper component needed for transmission. RTBV is not mechanically transmissible. There is no evidence of seed transmission. Host range is limited to members of the Poaceae and Cyperaceae.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Tungrovirus
| Rice tungro bacilliform virus |
|
|
| Rice tungro bacilliform virus-Philippines | [X57924=NC_001914] | (RTBV-PH) |
Species name is in italic script; name of isolate is in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Tungrovirus but have not been approved as species
None reported.
Phylogenetic relationships within the family Caulimoviridae
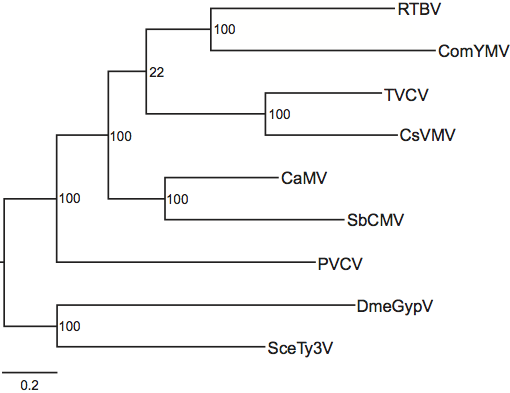
Phylogenetic relationships within the family have not yet been fully resolved and some of the relationships that are deduced depend on the method of analysis. However, several systematics studies have concluded that the genus Petuvirus is sister to all other genera in the Caulimoviridae and that the genera Badnavirus and Tungrovirus form a monophyletic group, as do the genera Soymovirus and Caulimovirus (Figure 3).
Similarity with other taxa
Members of the family Caulimoviridae have the conserved gag-pol replication core of all viral retroelements, suggesting a common ancestry. Phylogenetic analyses using conserved polymerase gene sequences suggest that the Caulimoviridae is sister to the Metaviridae. In the literature, members of the Caulimoviridae are frequently referred to as being plant-infecting pararetroviruses. The term “pararetrovirus” was coined to describe viral retroelements that do not integrate in the host genome as part of the replication cycle and which encapsidate dsDNA instead of ssRNA. However, the two “pararetrovirus” families, Hepadnaviridae and Caulimoviridae, are distantly related and a group containing these two families is polyphyletic.
Derivation of names
Badna: from bacilliform DNA viruses.
Caulimo: from cauliflower mosaic virus.
Cavemo: from cassava vein mottle virus
Petu: from petunia
Soymo: from soybean chlorotic mottle virus
Tungro: from rice tungro bacilliform virus
Further reading
Journals and books
Bousalem, M., Douzery, E. and Seal, S. (2008) Taxonomy, molecular phylogeny and evolution of plant reverse transcribing viruses (family Caulimoviridae) inferred from full-length genome and reverse transcriptase sequences. Arch. Virol., 153, 1085-1102.
Geering, A.D.W., Scharaschkin, T. and Teycheney, P.-Y. (2010). The classification and nomenclature of endogenous viruses of the family Caulimoviridae. Arch. Virol., 155, 123-131.
Haas, M., Bureau, M., Geldreich, A., Yot, P. and Keller, M. (2002). Cauliflower mosaic virus: still in the news. Mol. Plant Pathol., 3, 419-429.
Hoh, F., Uzest, M., Drucker, M., Plisson-Chastang, C., Bron, P., Blanc, S. and Dumas, C. (2010). Structural insights into the molecular mechanisms of Cauliflower mosaic virus transmission by its insect vector. J. Virol., 84, 4706-4713.
Hohn, T. and Richert-Poeggeler, K.R. (2006). Replication of plant pararetroviruses. In: Recent Advances in DNA Virus Replication (K.L. Hefferson, Ed.), Research Signpost 37/661, Kerala, India, pp. 289-319.
Plisson, C., Uzest, M., Drucker, M., Froissart, M., Dumas, C., Conway, J., Thomas, D., Blanc, S. and Bron, P. (2005). Structure of the mature P3-virus particle complex of cauliflower mosaic virus revealed by cryo-electron microscopy. J. Mol. Biol., 346, 267-277.
Pooggin, M.M., Fütterer, J., Skryabin, K.G. and Hohn, T. (1999). A short open reading frame terminating in front of a stable hairpin is the conserved feature in pregenomic RNA leaders of plant pararetroviruses. J. Gen. Virol., 80, 2217-2228.
Ryabova, L.A., Pooggin, M.M. and Hohn, T. (2006). Translation reinitiation and leaky scanning in plant viruses. Virus Res., 119, 52-62.
Stavolone, L., Herzog, E., Leclerc, D. and Hohn, T. (2001). Tetramerization is a conserved feature of the virion-associated protein in plant pararetroviruses. J. Virol., 75, 7739-7743.
Teycheney, P.-Y. and Geering, A.D.W. (2011). Endogenous viral sequences in plant genomes. In: Recent Advances in Plant Virology (C. Caranta, M.A. Aranda, M. Tepfer and J.J. Lopez-Moya, Eds.), Academic Press, Caister, pp. 347-366.
Websites
Descriptions of Plant Viruses Online: http://www.dpvweb.net/index.php
Contributed by
Geering, A.D.W. and Hull, R.
Figures
Figure 1 (Top left) Reconstruction of the surface structure of a cauliflower mosaic virus particle showing T=7 symmetry. (Top right) Cutaway surface reconstruction showing multilayer structure.
(From Cheng et al. (1992). Virology, 186, 655668). (Bottom) Negative contrast electron micrograph of particles of Commelina yellow mottle virus, stained with 2% sodium phosphotungstate, pH 7.0. The bar represents 10 nm.
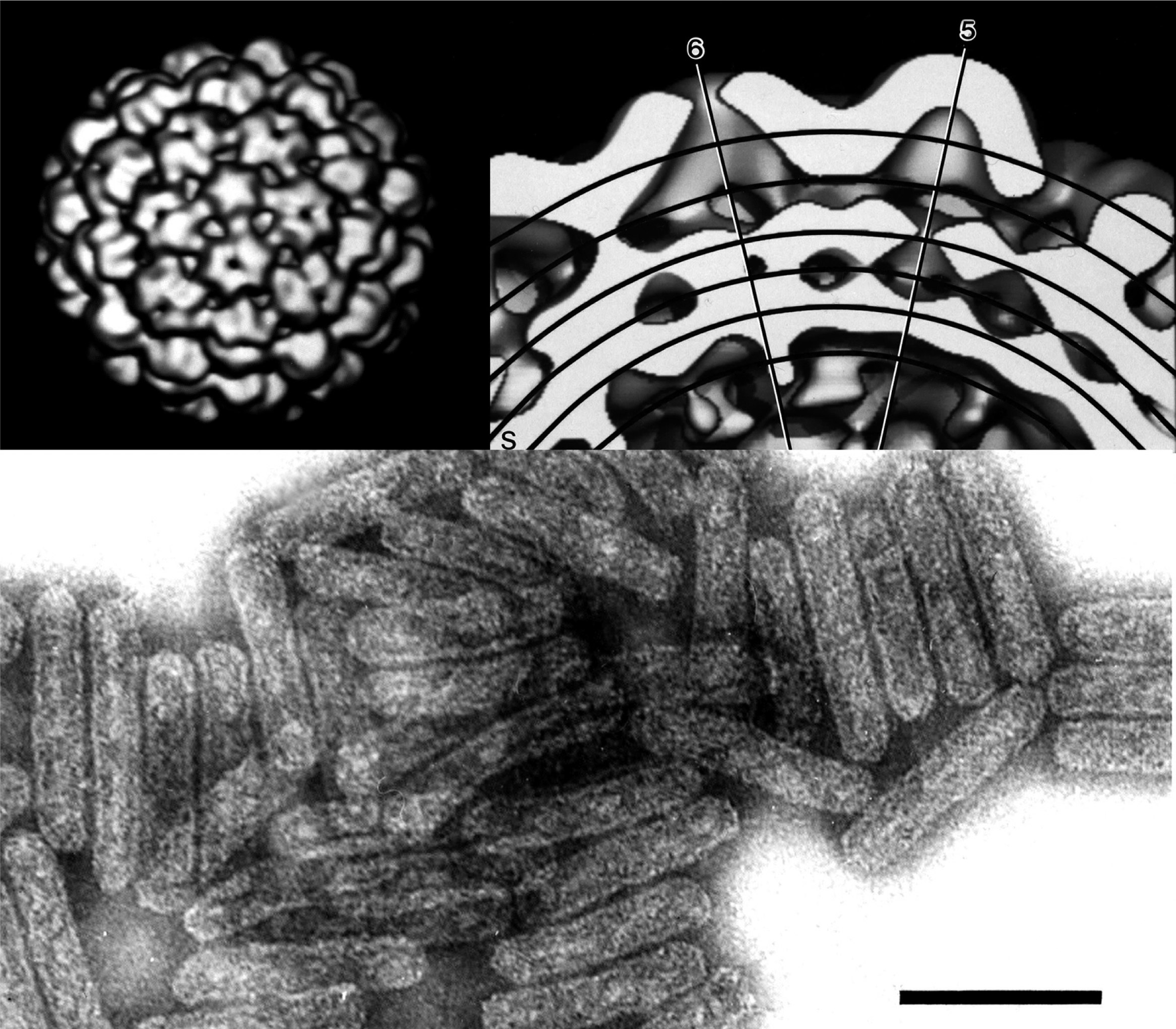
Figure 2 Comparison of the genome organizations of the different genera in the family Caulimoviridae. For all viruses except soybean mosaic virus (SbMV), the linearized maps begin at the 5 nucleotide of the minus-strand primer-binding site (designated by a black diamond). For ease of comparison, the start point of the SbCMV genome is the beginning of ORF7. Light grey boxes mark open reading frames and regions within each ORF that are coloured are conserved protein domains as listed in the Pfam database (http://pfam.sanger.ac.uk/): blue is the viral movement protein domain (PF01107), corresponding to L43E243 of the cauliflower mosaic virus (CaMV) ORF1 protein; red is the retropepsin (pepsin-like aspartic protease) domain (CD00303), corresponding to K36Q120 of the CaMV ORF5 protein; orange is the reverse transcriptase domain (CD01647), corresponding to K273G449 of the CaMV ORF5 protein; and yellow is the RNase H1 domain (CD06222), corresponding to I547E673 of the CaMV ORF5 protein. Additionally, coiled-coil motifs that are characteristic of the virion-associated protein are marked purple, the conserved C-terminus of the coat protein, corresponding to L261N429 of the CaMV ORF4 protein, is marked green, the translation transactivator active site is marked black and the position of RNA promoters is marked with an arrow.

Figure 3 Evolutionary relationships within the family Caulimoviridae based on comparison of the polymerase polyprotein gene. To obtain the nucleotide sequence alignment, amino acid sequences homologous to K36E673 of the cauliflower mosaic virus (CaMV) P5 protein (GenBank accession NP_056728) were first aligned with ClustalX and this alignment then used to generate a DNA alignment using the program Tranalign. Poorly aligned and highly variable regions in the alignment were then removed using the program Gblocks. Maximum likelihood analysis was done using the program RAxML and the GTR+ model of evolution to calculate the final tree topology. The dataset was partitioned into aspartic protease, reverse transcriptase and RNase H1 domains (see Figure 2 for domain boundaries), as well as tether regions between the domains. 1000 maximum likelihood bootstrap replicates were run (bootstrap values shown in nodes of branches) using the GTR+CAT approximation of GTR+. Drosophila melanogaster Gypsy virus (DmeGypV; genus Errantivirus) and Saccharomyces cerevisiae Ty3 virus (SceTy3V; genus Metavirus) were included in the analyses as outgroups. Other acronyms are ComYMV (Commelina yellow mottle virus), CsVMV (cassava vein mosaic virus), PVCV (petunia vein clearing virus), SbCMV (soybean chlorotic mottle virus), RTBV (rice tungro bacilliform virus) and TVCV (tobacco vein clearing virus).