Family: Virgaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Distinguishing features
The family Virgaviridae consists of plant viruses with rod-shaped virions, a single stranded RNA genome with a 3′-terminal tRNA-like structure and an alpha-like replication protein.
Virion properties
Morphology
The non-enveloped, rod-shaped particles are helically constructed with a pitch of 2.3 to 2.5 nm and an axial canal. They are about 20 nm in diameter, with predominant lengths that depend upon the genus.
Physicochemical and physical properties
The S20,w values range from 194 to 306 for large particles that encode the replication protein and 125 to 245 for smaller particles. Particles are stable at higher temperatures (60–90 °C) with the exception of pomoviruses, which lose infectivity at room temperature within a few hours.
Nucleic acid
The genome consists of positive sense ssRNA with 5′-cap (m7GpppG) and a 3′-terminal tRNA-like structure. The number of genome components depends upon the genus.
Proteins
The capsid comprises multiple copies of a single polypeptide of about 17–24 kDa, depending upon the genus. Some viruses encode an additional capsid protein (CP) produced by suppression of the CP gene stop codon to produce a larger readthrough (RT) protein of variable mass called the minor CP or CP-RT.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The largest (and 5′-most) ORF is an alpha-like replication protein with conserved methyltransferase (Mtr) and helicase (Hel) domains. This is believed to be translated directly from the genomic RNA. In all genera except Hordeivirus, the RNA-dependent RNA polymerase (RdRp) is expressed as the C-terminal part of this protein by readthrough of a leaky stop codon. Downstream genes are expressed from subgenomic RNAs, some of which may be bicistronic. In some genera, the viruses have a single cell-to-cell movement protein (MP) of the “30K” superfamily, while in other genera there is a triple gene block (TGB). There are differences in the number of genomic RNAs (1, 2 or 3 depending on the genus). Replication is cytoplasmic.
Antigenic properties
Virions are moderately to strongly antigenic.
Biological properties
Biologically, the viruses are fairly diverse. They have been reported from a wide range of herbaceous and mono- and dicotyledonous plant species, but the host range of individual members is usually limited. All members can be transmitted experimentally by mechanical inoculation, and for those in the genus Tobamovirus, this is the only known means of transmission. In some genera, transmission is by soil-borne vectors, while members of the genus Hordeivirus are transmitted through pollen and seed.
Genus demarcation criteria in the family
Genera are distinguished by the number of genomic RNAs, various features of genome organization, the type of cell-to-cell movement protein and the natural mode of transmission. These are summarized in Table 1.
Table 1 Distinguishing properties of genera in the family Virgaviridae
| Genus | RNAs | RdRPa | MPb | CPc | 3′ structured | Transmission |
| Furovirus | 2 | RT | “30K” | 19K+RT | t-RNAVal | “fungus” |
| Hordeivirus | 3 | Separate | TGB | 22K | t-RNATyr | seed |
| Pecluvirus | 2 | RT | TGB | 23K | t-RNAVal | “fungus” + seed |
| Pomovirus | 3 | RT | TGB | 20K+RT | t-RNAVal | “fungus” |
| Tobamovirus | 1 | RT | “30K” | 17–18K | t-RNAHis | mechanical |
| Tobravirus | 2 | RT | “30K” | 22–24K | t-RNA- | nematode |
a Relation of RdRp to the replication protein (methyltransferase, helicase); RT, in a readthrough domain at the C-terminus.
b MP, Movement protein either of the “30K” superfamily or a triple gene block (TGB).
c CP, Coat protein size in kDa (with indication of RT, a readthrough domain at the C-terminus if present).
d t-RNAVal/Tyr/His/-, t-RNA like structure accepting valine, tyrosine, histidine or not aminoacylated respectively.
Genus Furovirus
Type species Soil-borne wheat mosaic virus
Distinguishing features
Furoviruses have a bipartite genome, a “30K”-like cell-to-cell movement protein and are transmitted by root-infecting vectors in the family Plasmodiphorales, once described as fungi but now classified as Cercozoa.
Virion properties
Morphology
Virions are non-enveloped hollow rods, which have helical symmetry. Virions are about 20 nm in diameter, with predominant lengths of 140–160 nm and 260–300 nm. The length distribution of the soil-borne wheat mosaic virus (SBWMV) short particles is broad, 80–160 nm, due to the presence of deletion mutants in some cultures (Figure 1).
Physicochemical and physical properties
Virions sediment as two (or three) components; for SBWMV the S20,w values are 220–230S (long particles) and 170–225S (short particles), and 126–177S (deletion mutants). SBWMV loses infectivity in extracts of wheat kept at 60–65 °C for 10 min.
Nucleic acid
Complete or almost complete nt sequences are available for all five species in the genus. The genome is bipartite, linear, positive sense ssRNA. RNA-1 is about 6–7 kb and RNA-2 about 3.5–3.6 kb. The RNA molecules of SBWMV have a 5′ cap (m7GpppG) and in all of the species where the complete sequences have been determined there is a 3′-terminal tRNA-like structure with a putative anti-codon for valine. The 3′ terminus of SBWMV RNA was shown experimentally to accept valine.
Proteins
The capsid comprises multiple copies of a single polypeptide of about 19–20.5 kDa. The CPs of SBWMV, Chinese wheat mosaic virus (CWMV), soil-borne cereal mosaic virus (SBCMV) and oat golden stripe virus (OGSV) comprise 176 aa with 76–82% aa homologies; they share only 46% homology with that of sorghum chlorotic spot virus (SrCSV). The CP gene terminates in a leaky (UGA) stop codon that can be suppressed to produce a read-through protein (ca. 85 kDa), which is thought to be involved in natural transmission by the plasmodiophorid vector. In addition to replicase proteins the furoviruses encode a single MP (ca. 37 kDa) and a cysteine rich protein (ca. 18 kDa) that is probably a suppressor of gene silencing.
Genome organization and replication
Genome organization and structure is conserved between species but there are substantial differences in the nt sequences. SBWMV RNA-1 encodes a 150 kDa protein, a 209 kDa readthrough product and a 37 kDa protein (Figure 2). The 150 kDa protein contains Mtr and NTP-binding Hel motifs and the readthrough protein, in addition, contains RNA polymerase motifs, indicating that these proteins are involved in replication. The 37 kDa protein belongs to the “30K”-like cell-to-cell movement protein superfamily and is thought to be involved in virus movement as it shares partial sequence similarity to the MPs of dianthoviruses. RNA-2 encodes the CP (19 kDa), the sequence of which terminates in a UGA codon that can be suppressed to give a readthrough product of 84 kDa. A 25 kDa polypeptide is initiated from a CUG codon upstream of the CP AUG. An ORF towards the 3′ end of the RNA-2 encodes a 19 kDa protein that contains seven conserved cysteine residues. Products corresponding to the 37 kDa protein and the cysteine-rich 19 kDa protein were not found in in vitro transcription/translation experiments, and these proteins are thought to be expressed from sgRNAs. Spontaneous deletions in the CP readthrough domain occur on successive passage by manual inoculation, and in field isolates in older infected plants.
Antigenic properties
Virions are immunogenic and the five virus species can be distinguished serologically.
Biological properties
Host range
The natural host ranges of furoviruses are narrow and confined to species within the Graminae. SBWMV induces green or yellow mosaic and stunting in winter wheat (Triticum aestivum) causing up to 80% yield loss in severely infected crops. It also may infect barley and rye. SBCMV infects mainly wheat and triticale in Western and Southern Europe and mainly rye in Central and North-Eastern Europe. Both viruses are (not readily) mechanically transmissible to Chenopodium quinoa. OGSV infects oats (Avena sativa) but failed to infect wheat when plants were grown in viruliferous soil. Mechanically it can be transmitted to some Nicotiana and Chenopodium species. SrCSV infects Sorghum bicolor. Mechanically it can be transmitted to a range of species including Chenopodium quinoa, C. amaranticolor, Nicotiana clevelandii, Arachis hypogaea, Zea mays and T. aestivum.
Transmission
The viruses are soil-borne, and Polymyxa graminis has been identified as a vector for SBWMV. Virions are thought to be carried within the motile zoospores. Soil containing the resting spores remains infectious for many years.
Geographical distribution
Furoviruses are found in temperate regions worldwide including the United States of America, Europe, China, Japan.
Cytopathic effects
Virions are found scattered, or in aggregates and inclusion bodies in the cytoplasm and vacuole. Inclusion bodies can be crystalline inclusions or comprise loose clusters of virus particles in association with masses of microtubules. Amorphous inclusion bodies can be seen in tissue sections by light microscopy.
Species demarcation criteria in the genus
The species within the genus Furovirus are presently mainly differentiated on the basis of the nt sequences of their RNAs and the deduced aa sequences of their putative gene products. RNA-1 sequences of these viruses share 58–74% identity and RNA-2 46–80% (see Figure 3). SBWMV, SBCMV, CWMV and OGSV can be discriminated also by reactivity with selected monoclonal and polyclonal antibodies. OGSV and SrCSV differ in host range to SBWMV, SBCMV and CWMV. Especially the latter three viruses have similar biological properties and genetic reassortants can be formed with RNA-1 and RNA-2 of SBWMV (Nebraska), SBWMV (Japan) and SBCMV. With the other viruses this possibility has not yet been checked.
List of species in the genus Furovirus
| Chinese wheat mosaic virus | ||
| Chinese wheat mosaic virus-China:Yantai | [AJ012005=NC_002359 + AJ012006=NC_002356] | (CWMV-YT) |
| Oat golden stripe virus | ||
| Oat golden stripe virus-UK | [AJ132578=NC_002358 + AJ132579=NC_002357] | (OGSV-UK) |
| Soil-borne cereal mosaic virus | ||
| (European wheat mosaic virus) |
|
|
| (Soil-borne rye mosaic virus) |
|
|
| Soil-borne cereal mosaic virus-France | [AJ132576=NC_002351 + AJ132577=NC_002330] | (SBCMV-FR) |
| Soil-borne wheat mosaic virus | ||
| Soil-borne wheat mosaic virus-USA:Nebraska | [L07937=NC_002041 + L07938=NC_002042] | (SBWMV-NE) |
| Sorghum chlorotic spot virus | ||
| Sorghum chlorotic spot virus-USA | [AB033691=NC_004014 + AB033692=NC_004015] | (SrCSV-USA) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Furovirus but have not been approved as species
| French barley mosaic virus | [AJ749657-8*] |
|
| Japanese soil-borne wheat mosaic virus | [AB033689-70] | (JSBWMV) |
* Sequences do not comprise the complete genome.
Genus Hordeivirus
Type species Barley stripe mosaic virus
Distinguishing features
Hordeiviruses have three genomic RNAs and a “triple gene block” set of cell-to-cell movement proteins. They differ from all other genera because the RNA-dependent RNA polymerase (RdRp) is encoded on a separate RNA (rather than by readthrough of a stop codon from an upstream replication protein).
Virion properties
Morphology
Virions are non-enveloped, elongated and rigid, about 20×110–150 nm in size; they are helically symmetrical with a pitch of 2.5 nm (Figure 4).
Physicochemical and physical properties
Barley stripe mosaic virus (BSMV) virions occur as heterodisperse sedimenting species with an S20,w of about 182–193S; other species have an S20,w of about 165–200S, depending on the virus. The BSMV isoelectric point is pH 4.5. Anionic detergents, added to purification buffers, increase virus yield by preventing particle aggregation. Thermal inactivation of infectivity occurs at 63–70 °C. Virions are stable and their survival in sap ranges from a few days to several weeks.
Nucleic acid
Virions normally contain three positive sense ssRNAs. The RNAs are designated α, β and γ, and their respective sizes are 3.8, 3.2 and 2.8 kb (BSMV-ND18 strain), 3.7, 3.1 and 2.6 kb (Lychnis ringspot virus, LRSV), and 3.9, 3.6 and 3.2 kb (Poa semilatent virus, PSLV). The sizes of the α and β RNAs are similar between different strains of BSMV, whilst RNAγ varies in size. The ND18 RNAγ is 2.8 kb, that of the type strain is 3.2 kb. This difference is due to a 266 nt duplication near the 5′ end of the RNA that produces a γa protein of 87 kDa in the type strain compared to a 74 kDa γa ND18 protein. The Argentine mild strain contains mixtures of RNAγ species of 3.2, 2.8 and 2.6 kb. The 3.2 kb molecule contains a duplication similar to that of the type strain and the 2.6 kb RNA encodes a defective polymerase. No extensive hybridization can be detected between RNAs of BSMV, LRSV and PSLV. Each RNA has m7GpppGUA at its 5′ end, and a highly conserved 238 nt (BSMV), 148 nt (LRSV), or 330 nt (PSLV) tRNA-like structure at the 3′ end. In the case of BSMV, this structure can be charged with tyrosine. In the BSMV and LRSV genomes, a poly(A) sequence that is variable in length separates the coding region from the tRNA-like structure; however, this sequence is not present in the PSLV genome. A close sequence similarity between the first 70 nt of RNAα and RNAγ of the CV17 strain of BSMV suggests that a natural recombination event has occurred between RNAα and RNAγ of this strain. A similar recombination appears to have occurred between the 5′-untranslated leaders of RNAα and RNAβ of LRSV. These results plus sequence duplications in RNAγ provide persuasive evidence that RNA recombination has had a substantial role in the evolution of hordeiviruses.
Proteins
The virion capsid is constructed from subunits of a single protein. The CP of all species is 22 kDa in size, yet the proteins differ in electrophoretic mobility.
Genome organization and replication
All three BSMV genomic RNAs are required for systemic infection of plants, but RNAs α and γ alone can infect protoplasts. The 5′- and 3′-NCR of each BSMV RNA are required for replication. The hordeivirus genome encodes seven proteins as illustrated for BSMV in Figure 2. RNAα is monocistronic and encodes the αa protein (130 kDa in BSMV, 129 kDa in LRSV and 131 kDa in PSLV) that functions as the helicase subunit of the viral replicase. The αa protein has two conserved sequence domains, an amino-terminal Mtr and a carboxy-terminal NTPase/Hel. The 5′-terminal RNAβ ORFs (βa) of all three viruses encode a 22 kDa CP. The BSMV CP, which is dispensable for systemic movement of the virus, is more closely related to the PSLV CP (55.2% identity) than to the LRSV CP (41.5% identity). An intergenic region separates a “triple gene block” (TGB) that encodes three nonstructural proteins, βb (TGB1), βc (TGB3) and βd (TGB2), in which the βd protein overlaps the other two genes. In BSMV, The βb protein is expressed from a 2,450 nt sgRNA, and the βc and βd proteins are expressed from a second bicistronic 960 nt sgRNA with βc being expressed via a leaky scanning mechanism. In BSMV, a minor 23 kDa translational readthrough extension of the βd protein, designated βd′, is present in plants. However, genetic experiments have not identified a function for βd′, so it appears to be dispensable for infection in all local lesion and systemic hosts tested. The BSMV sgRNAβ1 and sgRNAβ2 promoters reside between positions −29 to −2 and −32 to −17 relative to the transcription initiation sites, respectively, and the nt sequences preceding the transcription initiation sites of these sgRNAs are conserved in LRSV and PSLV. The βb protein (58 kDa in BSMV, 50 kDa in LRSV, and 63 kDa in PSLV) contains a conserved NTPase/Hel domain. The BSMV βb protein binds RNA, NTPs and exhibits ATPase and helicase activity in vitro. The βc (17 kDa in BSMV, and 18 kDa in LRSV and PSLV) and βd (14 kDa in BSMV and LRSV, and 18 kDa in PSLV) proteins are hydrophobic and membrane-associated. Each of the BSMV TGB proteins (βb, βc and βd) is required for virus cell-to-cell movement in plants. RNAγ is bicistronic and encodes the γa polymerase subunit of the viral replicase (74 kDa in the BSMV-ND18 strain, 71 kDa in LRSV, and 84 kDa in PSLV), and the cysteine-rich γb protein (17 kDa in BSMV, 16 kDa in LRSV, and 20 kDa in PSLV) (see Figure 5).
The γa protein is variable in size because of the approximately 250 nt RNAγ repeated sequence present in different strains of BSMV. The BSMV γb protein is expressed from a 737 nt sgRNA and is a pathogenicity determinant that is involved in regulating expression of genes encoded by RNAβ. The sgRNAγ promoter is between nt −21 to +2 relative to its transcription start site, and this sequence has similarity to sequences upstream of the γb proteins in PSLV and LRSV. The BSMV γb protein has both RNA binding and zinc binding ability, participates in homologous interactions, and may act as a suppressor of post-transcriptional gene silencing. Translation of a functional αa protein is required for replication of RNAα in cis, whilst replication of RNAβ is dependent on the presence of the βa and βb intergenic region, and RNAγ requires approximately 600 nt of the 5′-terminal region. The TGB proteins on RNAβ (b, c, d) are required for cell-to-cell and systemic movement in plants, but the CP and βd′ are dispensable. The γb protein is also dispensable in some genetic backgrounds. A mutation in the 5′-leader sequence of the γa ORF interfered with systemic infection of Nicotiana benthamiana, suggesting that modulation of γa expression can affect movement. Full-length dsRNAs corresponding to all viral genomic ssRNAs can be isolated from infected plants. Virus particles accumulate predominantly in the cytoplasm and also in nuclei. Infected barley plants develop pronounced enlargements of the plasmodesmata that contain the βb protein, and prominent peripheral vesicles appear in proplastids and chloroplasts. These vesicles may be the sites of replication because antibodies raised against poly(I):poly(C) have detected dsRNA in proplastids from infected barley root tips.
Antigenic properties
Hordeivirus particles are efficient immunogens. Member species are distantly related serologically with BSMV being more closely related to PSLV than to LRSV, which is in agreement with sequence analyses.
Biological properties
Host range
The native hosts of three viruses (ALBV, BSMV, PSLV) are grasses (family Gramineae); strains of LRSV occur naturally in dicotyledonous plants of the families Caryophyllaceae and Labiatae. Various strains of these viruses elicit local lesions in Chenopodium species and are able to establish systemic infections in a common host, Nicotiana benthamiana.
Transmission
BSMV and LRSV are efficiently seed-transmitted, and are transmitted less efficiently by pollen. Field spread from primary infection foci occurs efficiently by direct leaf contact. There are no known vectors for any members of the genus.
Geographic distribution
ALBV has been reported only from Great Britain; BSMV occurs world-wide wherever barley is grown; LRSV (mentha strain) has been isolated in Hungary, and the type strain which is highly seed-transmissible in the family Caryophyllaceae, was initially discovered in California from seed of Lychnis divaricata introduced from Europe. PSLV has been recovered from Poa palustris isolated from two locations in Western Canada.
Species demarcation criteria in the genus
Species differ in host range and are phylogenetically distinct in the genes studied. Precise molecular discrimination criteria have not been established because few sequences have been determined except for the type member.
List of species in the genus Hordeivirus
| Anthoxanthum latent blanching virus |
|
|
| Anthoxanthum latent blanching virus-UK:Aberystwyth |
| (ALBV-ABER) |
| Barley stripe mosaic virus |
|
|
| Barley stripe mosaic virus-Type | [J04342=NC_003469+ X03854=NC_003481+ M16576=NC_003478] | (BSMV-Type) |
| Lychnis ringspot virus |
|
|
| Lychnis ringspot virus-USA:California | [Z46351*+Z46353*] | (LRSV-CAL) |
| Poa semilatent virus |
|
|
| Poa semilatent virus-Canada | [M81486*+M81487*] | (PSLV-CAN) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Hordeivirus but have not been approved as species
None reported.
Genus Pecluvirus
Type species Peanut clump virus
Distinguishing features
Pecluviruses have a bipartite genome, a “triple gene block” set of cell-to-cell movement proteins and are transmitted by root-infecting vectors in the family Plasmodiphorales, once described as fungi but now classified as Cercozoa.
Virion properties
Morphology
Virions are rod-shaped of about 21 nm in diameter and of two predominant lengths of 190 and 245 nm (Figure 6). The length distribution of the short particles is broad and in some preparations an additional class of 160 nm is recognizable. Virions have helical symmetry with a pitch of 2.6 nm.
Physicochemical and physical properties
Virions sediment as two major components with S20,w of 183S and 224S. Buoyant density in CsCl is 1.32 g cm−3. Virion isoelectric point is pH 6.45. Thermal inactivation of infectivity occurs at 64 °C. Virions are stable in frozen leaves.
Nucleic acid
The genome consists of two molecules of linear positive sense ssRNA; RNA-1 of about 5,900 nt and RNA-2 of about 4,500 nt. RNAs are thought to have a 5′-cap structure but this has not been confirmed. The 3′ ends of the RNAs can fold into a tRNA-like structures and are not polyadenylated.
Proteins
The virion CP subunits are 23 kDa.
Genome organization and replication
RNA-1 contains two ORFs (Figure 7). The 5′ ORF encodes a 131 kDa protein, and suppression of a termination codon results in the synthesis of a readthrough protein of 191 kDa. The 3′ ORF encodes a 15 kDa protein. The proteins of 131 and 191 kDa contain NTP-binding, Hel and RNA polymerase motifs that make a putative replication complex. For peanut clump virus (PCV), these proteins are respectively 88%, 95% and 75% similar to the products of an isolate of one serotype of Indian peanut clump virus (IPCV). The 15 kDa protein is translated from a sgRNA. It is a suppressor of post-transcriptional gene silencing and is targeted to peroxisomes or related punctate bodies during infection. RNA-2 contains five ORFs: the ORF near the 5′ end encodes the CP, the second ORF which, in PCV RNA-1, overlaps the first ORF by 2 nts encodes a 39 kDa protein. This protein is expressed by leaky scanning and is thought to be involved in the transmission of PCV by its fungus vector. Further downstream, separated by a 135 nt intergenic region, is a triple gene block sequence that codes for proteins of 51, 14 and 17 kDa that are thought to be involved in the movement of virus from cell to cell. The proteins are probably expressed via sgRNAs, but these have not been clearly identified. The 3′-NCRs for PCV are 298 nt for RNA-1 and 275 nt for RNA-2; the last 96 nt are identical in both RNAs. The NCRs differ in size among isolates from the different serotypes of IPCV.
The two RNAs are required for systemic invasion of plants but RNA-1 is able to replicate in absence of RNA-2 in protoplasts. The virus is found in the cells of roots, stems and leaves of systemically infected plants.
Antigenic properties
The virus is highly immunogenic. There is a great serological variability among isolates of PCV. IPCV isolates fall into one of three very distinct serotypes: IPCV-H, IPCV-L, IPCV-T. All are serologically distinct from PCV.
Biological properties
Host range
The natural host first reported was Arachis hypogea (groundnut, Leguminosae). Disease symptoms are stunting – mottle – mosaic – chlorotic ringspot. PCV infects Sorghum arundinaceum, usually symptomlessly. IPCV infects a number of cereal crops and graminaceous weeds, some symptomlessly and others to induce stunting. The experimental host range is wide and includes species of Aizoaceae, Amaranthaceae, Chenopodiaceae, Cucurbitaceae, Graminae, Leguminosae, Scrophulariaceae and Solanaceae. Nicotiana benthamiana and Phaseolus vulgaris are experimental propagation hosts, Chenopodium amaranticolor and Chenopodium quinoa are local lesions hosts.
Transmission
The virus is transmitted naturally by Polymyxa graminis or by seed (in groundnuts). It is mechanically transmissible.
Geographical distribution
PCV spreads in West Africa (Bénin, Burkina Faso, Congo, Côte d’Ivoire, Mali, Niger, Senegal and Pakistan). IPCV is widely distributed in India and Pakistan. A soil type favorable to the vector is a prerequisite for virus to cause disease.
Species demarcation criteria in the genus
The species are distinguished by different reactions with particular antisera (heterologous reactions are weak or undetectable). Also, PCV occurs only in Africa whereas IPCV occurs in the Indian subcontinent. However, isolates of IPCV can be readily assigned to one of three serotypes as protein preparations made from particles of each serotype barely react with heterologous antisera in immunoblotting tests. Isolates of PCV are also heterogeneous in their reactions with a panel of monoclonal antibodies. Moreover, several of the proteins encoded by genes in RNA of the different serotypes of IPCV differ in sequence from corresponding proteins of other IPCV serotypes by about as much as each differs from the corresponding protein of one isolate of PCV.
List of species in the genus Pecluvirus
| Indian peanut clump virus | ||
| Indian peanut clump virus-Hyderabad serotype | [X99149=NC_004729 + AF447397=NC_004730] | (IPCV-H) |
| Peanut clump virus | ||
| Peanut clump virus-87/TGTA2 | [X78602=NC_003672 + L07269=NC_003668] | (PCV-87/TGTA2) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Pecluvirus but have not been approved as species
None reported.
Genus Pomovirus
Type species Potato mop-top virus
Distinguishing features
Pomoviruses have three genomic RNAs, a “triple gene block” set of cell-to-cell movement proteins and are transmitted by root-infecting vectors in the family Plasmodiophorales, once described as fungi but now classified as Cercozoa.
Virion properties
Morphology
The non-enveloped, rod-shaped particles are helically constructed with a pitch of 2.4 to 2.5 nm and an axial canal (Figure 8). They have predominant lengths of about 65–80, 150–160 and 290–310 nm and diameters of 18–20 nm. Crude extracts of plants infected with beet soil-borne virus (BSBV), beet virus Q (BVQ) and potato mop-top virus (PMTV) contain characteristic small bundles of a few side-by-side aggregated particles in addition to singly dispersed particles.
Physicochemical and physical properties
Virions sediment as three components with S20,w of about 125S, 170S and 230S, respectively. In sap at room temperature, most of the infectivity is lost within a few hours.
Nucleic acid
Virions contain three molecules of linear positive sense ssRNA of about 6, 3–3.5 and 2.5–3 kb, respectively. The sequence has been determined for all three RNA species of BSBV, BVQ, PMTV and broad bean necrosis virus (BBNV). The RNAs are probably capped at the 5′ end; their 3′ ends can be folded into tRNA-like structures that are preceded by a long hairpin-like structure and an upstream pseudoknot domain. The tRNA-like structures of pomoviruses like those of tymoviruses contain an anticodon for valine and are capable of high-efficiency valylation.
Proteins
The major capsid protein (CP) species is 20 kDa in size. It is not needed for systemic infection. The CP readthrough protein may be detected in some PMTV particles near one extremity by means of immunogold labeling. Sequences in the CP readthrough protein are necessary for the transmission of PMTV by Spongospora subterranea. Yeast two-hybrid experiments revealed that the CP readthrough protein interacts with the triple gene block protein movement protein TGB1. In this system, TGB proteins show self interactions and TGB2 and TGB3 interact with each other. TGB2 and TGB3 are membrane-associated and TGB2 binds ssRNA in a sequence nonspecific manner. It has been suggested that they may form a complex with PMTV RNA that is translocated and localized to the plasmodesmata by TGB3.
Genome organization and replication
RNA-1 of PMTV has an ORF for a 148 kDa protein and a 206 kDa readthrough protein that are presumably involved in replication. The shorter ORF is terminated by an apparently suppressible UGA stop codon (Figure 9). Proteins of similar sizes are encoded on RNA-1 of BVQ, BBNV and BSBV. The smaller protein contains a Mtr motif in its N-terminal part and a Hel motif in its C-terminal part; the motifs for RdRp are found in the C-terminal part of the readthrough protein (Figure 9). The two proteins contain other highly conserved domains of unknown function in their N- and C-terminal parts, but their central regions (designated as “variable” in Figure 9) are highly specific for each virus. RNA-CP (in PMTV) and the second largest RNA (RNA 2 of the other pomoviruses) contains the CP gene, which terminates with a suppressible UAG stop codon and then continues in frame to form a CP readthrough protein gene that varies considerably in size between different pomoviruses, possibly because it readily undergoes internal deletions, and large deletions have been found in both natural and laboratory isolates of PMTV. PMTV RNA-CP was therefore originally designated as RNA-3. PMTV RNA-2 contains a gene for a cysteine-rich protein and a gene encoding a predicted 6 kDa glycine-rich protein in BBNV neither of which are found on the RNAs of BSBV and BVQ. A triple gene block (TGB) coding for proteins involved in viral movement is found on RNAs-3. TGB1 also contains Hel motifs. The sequences of the C-terminal part of TGB1, of the entire TGB2 and of the N-terminal part of TGB3 are highly conserved among pomoviruses. The replication mechanisms are unknown.
Antigenic properties
Virions are moderately antigenic. Distant serological relationships have been found between the particles of BSBV and BVQ but not between those of the two beet viruses and PMTV. This is probably due to the fact that PMTV CP has ten extra amino acids on its immunodominant N-terminus that are missing in the two beet viruses. A conserved sequence EDSALNVAHQL is found in the CPs of PMTV, BSBV and BVQ. It contains an epitope for which the monoclonal antibody SCR 70 is specific and which is only detectable by Western blotting after disruption of the particles. Other epitopes are either exposed along the entire particle length, e.g. the immunodominant N-terminus, or are accessible only on one extremity (Figure 8). PMTV and BBNV show distant serological relationships to tobamoviruses.
Biological properties
Host range
The natural host range of pomoviruses is very narrow; only dicotyledonous hosts have been described.
Transmission
Pomoviruses are transmitted by soil. Spongospora subterranea and Polymyxa betae have been identified as vectors for PMTV and BSBV, respectively. The viruses are also transmissible mechanically.
Geographical distribution
Countries with temperate climate.
Cytopathic effects
PMTV-infected cells contain in the cytoplasm virions aggregated in sheaves. Infections by BSBV and BVQ induce voluminous cytoplasmic inclusions which consist of hypertrophied endoplasmic reticulum, convoluted membrane accumulations, numerous small virion bundles and rarely compact virus aggregates.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Differences in host range
- Effects in infected tissue: different inclusion body morphology
- Transmission: different vector species
- Serology: virions are distantly related serologically
- Genome: different numbers of genome components (presence or absence of a gene for a cysteine-rich protein)
- Sequence: less than about 80% identical over whole sequence
- Sequence: less than about 90% identical in CP amino acid sequence
List of species in the genus Pomovirus
| Beet soil-borne virus | ||
| Beet soil-borne virus-Ahlum | [Z97873=NC_003520 + U64512=NC_003518 + Z66493=NC_003519] | (BSBV-Ahlum) |
| Beet virus Q | ||
| Beet virus Q-Germany | [AJ223596=NC_003510 + AJ223597=NC_003511 + AJ223598=NC_003512] | (BVQ-DE) |
| Broad bean necrosis virus | ||
| Broad bean necrosis virus-Japan | [D86636=NC_004423 + D86637=NC_004424 + D86638=NC_004425] | (BBNV-JP) |
| Potato mop-top virus | ||
| Potato mop-top virus-Sweden:Halland | [AJ238607=NC_003723 + AJ243719=NC_003724 + AJ277556=NC_003725] | (PMTV-Sw) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Pomovirus but have not been approved as species
None reported.
Genus Tobamovirus
Type species Tobacco mosaic virus
Distinguishing features
Tobamoviruses are the only members of the family to have an undivided genome. They have a “30K”-like cell-to-cell movement protein, are not vector-transmissible and when seed transmitted the embryo is not affected. It is easily the largest genus in the family for numbers of species.
Virion properties
Morphology
Virions are 18 nm in diameter and have a predominant length of 300–310 nm (Figure 10). Shorter virions produced by the encapsidation of sgRNA are a minor component of the virion population, although at least two species produce an abundant short virion 32–34 nm in length. Virions often form large crystalline arrays visible by light microscopy.
Physicochemical and physical properties
Virion Mr is 40×106. Buoyant density in CsCl is 1.325 g cm−3. S20,w is 194S. Tobamoviruses have thermal inactivation points of 90 °C and survive in sap for many years.
Nucleic acid
The genome is 6.3–6.6 kb in size. An approximately 70 nt long 5′-untranslated sequence contains many AAC repeats and few or no G residues. The 0.2–0.4 kb 3′-UTR contains sequences that can be folded into pseudoknots followed by 3′-terminal sequences that can be folded into a tRNA-like, amino acid-accepting structure. SgRNAs also contain a 5′-terminal cap and 3′-tRNA-like structure. The origin of assembly for encapsidation is usually located within the ORF for the MP, but within the ORF for the CP in the studied isolates of at least two species, Cucumber green mottle mosaic virus and Sunn hemp mosaic virus.
Proteins
Virions contain a single structural protein (17–18 kDa). Two nonstructural proteins are expressed directly from the genomic RNA: a 124–132 kDa protein terminated by an amber (UAG) stop codon and a 181–189 kDa protein produced by readthrough of this stop codon, both of which are required for efficient replication. A third nonstructural protein (28–31 kDa) is required for cell-to-cell and long-distance movement and belongs to the “30K”-like cell-to-cell movement proteins. The MP is associated with plasmodesmata and has single-stranded nucleic acid binding activity in vitro. The CP is not required for cell-to-cell movement, but has a role in vascular tissue dependent virus accumulation. The replication proteins have also been implicated in virus movement. The MP and CP are expressed from individual 3′-co-terminal sgRNAs. The MP is expressed early during infection, whereas the CP is expressed later, and at higher levels. The MP and CP are not required for replication in single cells. The N-terminal one-third of the 124–132 kDa protein has similarity with methyltransferase/guanylyl transferases whereas the C-terminal one-third of the 124–132 kDa protein has similarity with RNA helicases (including an NTP-binding motif). The readthrough domain of the 181–189 kDa protein has motifs common to RdRps.
Genome organization and replication
The single genomic RNA encodes at least four proteins. The 124–132 kDa and 181–189 kDa replication proteins are translated directly from the genomic RNA. The 124–132 kDa replication protein contains the Mtr and Hel domains. The 181–189 kDa replication protein that also contains the polymerase domain is synthesized by occasional readthrough of the leaky termination codon of the 124–132 kDa ORF. The 181–189 kDa replication protein is the only protein required for replication in single cells, although the 124–132 kDa replication protein is also required for efficient replication. The next ORFs encode the 28–31 kDa MP and 17–18 kDa CP, which are translated from their respective 3′ co-terminal sgRNAs, both of which contain a 5′ cap (Figure 11). In some species, the MP ORF overlaps both of the 181–189 kDa protein and CP ORFs, and in others does not overlap either ORF or overlaps one of the ORFs.
Antigenic properties
The virions act as strong immunogens. Different species can be identified by intragel cross-absorption immunodiffusion tests using polyclonal antisera or by ELISA using monoclonal antibodies. Antigenic distances between individual species expressed as serological differentiation indices are correlated with the degree of sequence difference in their CPs.
Biological properties
Host range
Most species have moderate to wide host ranges under experimental conditions, although in nature host ranges are usually quite narrow. The viruses are found in all parts of host plants.
Transmission
Transmission occurs without the help of vectors by contact between plants and sometimes by seed, although this occurs in the absence of infection of the embryo.
Geographical distribution
Members of the genus are found throughout the world.
Species demarcation criteria in the genus
Many tobamoviruses that were historically designated as strains of tobacco mosaic virus are now defined as separate species based on nucleotide sequence data.
The criteria demarcating species in the genus are:
- Sequence similarity: less than 10% overall nt sequence difference is considered to characterize strains of the same species, although most of the sequenced species have considerably less than 90% sequence identity
- Host range: however many of these viruses have wider and more overlapping host ranges in experimental rather than natural situations
- Antigenic relationships between the CPs
List of species in the genus Tobamovirus
| Brugmansia mild mottle virus | ||
| Brugmansia mild mottle virus-2373 | [AM398436=NC_010944] | (BrMMV-2373) |
| Cucumber fruit mottle mosaic virus | ||
| Cucumber fruit mottle mosaic virus-Israel | [AF321057=NC_002633] | (CFMMV-IS) |
| Cucumber green mottle mosaic virus | ||
| Cucumber green mottle mosaic virus-SH | [D12505=NC_001801] | (CGMMV-SH) |
| Frangipani mosaic virus | ||
| Frangipani mosaic virus-China:Hainan | [AF165884*] | (FrMV-CN) |
| Hibiscus latent Fort Pierce virus | ||
| Hibiscus latent Fort Pierce virus-USA:Florida | [FJ196834*] | (HLFPV-FLA) |
| Hibiscus latent Singapore virus | ||
| Hibiscus latent Singapore virus-Singapore | [AF395898=NC_008310] | (HLSV-SIN) |
| Kyuri green mottle mosaic virus | ||
| Kyuri green mottle mosaic virus-C1 | [AJ295948=NC_003610] | (KGMMV-C1) |
| Obuda pepper virus | ||
| Obuda pepper virus-TMV-Ob | [D13438=NC_003852] | (ObPV-TMVOb) |
| Odontoglossum ringspot virus | ||
| Odontoglossum ringspot virus-Korea | [X82130=NC_001728] | (ORSV-KOR) |
| Paprika mild mottle virus | ||
| Paprika mild mottle virus-Japan | [AB089381=NC_004106] | (PaMMV-JP) |
| Pepper mild mottle virus | ||
| Pepper mild mottle virus-S | [M81413=NC_003630] | (PMMoV-S) |
| Rehmannia mosaic virus | ||
| Rehmannia mosaic virus-Henan | [EF375551=NC_009041] | (RheMV-HN) |
| Ribgrass mosaic virus | ||
| Ribgrass mosaic virus-Stubbs | [U69271*] | (RMV-Stubbs) |
| Sammons's Opuntia virus | ||
| Sammons’s Opuntia virus-USA:Arizona |
| (SOV-AZ) |
| Streptocarpus flower break virus | ||
| Streptocarpus flower break virus-Germany | [AM040955=NC_008365] | (SFBV-DE) |
| Sunn-hemp mosaic virus | ||
| Sunn-hemp mosaic virus-India | [U47034*]+[J02413*] | (SHMV-IN) |
| Tobacco latent virus | ||
| Tobacco latent virus-Nigeria | [AY137775*] | (TLV-NIG) |
| Tobacco mild green mosaic virus | ||
| Tobacco mild green mosaic virus-Canary Islands | [M34077=NC_001556] | (TMGMV-CY) |
| Tobacco mosaic virus | ||
| Tobacco mosaic virus-variant 1 | [V01408=NC_001367] | (TMV-1) |
| Tomato mosaic virus | ||
| Tomato mosaic virus-Australia: Queensland | [AF332868=NC_002692] | (ToMV-QLD) |
| Turnip vein-clearing virus | ||
| Turnip vein-clearing virus-OSU | [U03387=NC_001873] | (TVCV-OSU) |
| Ullucus mild mottle virus | ||
| Ullucus mild mottle virus-Peru |
| (UMMV-Peru) |
| Wasabi mottle virus | ||
| Wasabi mottle virus-Shizuoka | [AB017503=NC_003355] | (WMoV-Shizuoka) |
| Youcai mosaic virus | ||
| Youcai mosaic virus-OSRMV | [U30944=NC_004422] | (YMoV-OSRMV) |
| Zucchini green mottle mosaic virus | ||
| Zucchini green mottle mosaic virus-Type strain K | [AJ295949=NC_003878] | (ZGMMV-K) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Tobamovirus but have not been approved as species
| Abutilon yellow mosaic virus | [EU559678*] | (AbYMV) |
| Bell pepper mottle virus | [DQ355023=NC_009642] | (BPMoV) |
| Cactus mild mottle virus | [EU043335=NC_011803] | (CMMoV) |
| Cucumber mottle virus | [AB261167=NC_008614] | (CuMoV) |
| Maracuja mosaic virus | [DQ356949=NC_008716] | (MarMV) |
| Tropical soda apple mosaic virus | [AY956381-2*] | (TSAMV) |
* Sequences do not comprise the complete genome.
Genus Tobravirus
Type species Tobacco rattle virus
Distinguishing features
Tobraviruses have a bipartite genome, a “30K”-like cell-to-cell movement protein and are transmitted by trichodorid nematodes.
Virion properties
Morphology
Virions are tubular particles with no envelope (Figure 12). They are of two predominant lengths, (L) 180–215 nm and (S) ranging from 46 to 115 nm, depending on the isolate. Many strains produce in addition small amounts of shorter particles. The particle diameter is 21.3–23.1 nm by electron microscopy or 20.5–22.5 nm by X-ray diffraction, and there is a central canal 4–5 nm in diameter. Virions have helical symmetry with a pitch of 2.5 nm; the number of subunits per turn has been variously estimated as 25 or 32.
Physicochemical and physical properties
Virion Mr is 48–50×106 (L particles) and 11–29×106 (S particles). Buoyant density in CsCl is 1.306–1.324 g cm−3. S20,W is 286–306S (L particles) and 155–245S (S particles). Virions are stable over a wide range of pH and ionic conditions and are resistant to many organic solvents, but are sensitive to treatment with EDTA. In N. clevelandii sap, the thermal inactivation point (10 min) of M-type isolates is 80–85 °C.
Nucleic acid
The genome consists of two molecules of linear positive sense ssRNA; RNA-1 is about 6.8 kb and RNA-2 ranges from 1.8 kb to about 4.5 kb in size (varying in different isolates). The 5′ terminus is capped with the structure m7G5′ppp5′Ap. There is no genome-linked protein or poly(A) tract. The 3′ terminus can adopt a tRNA-like structure that can be adenylated but not aminoacylated.
Proteins
Virions contain a single structural protein of 22–24 kDa. RNA-1 codes for four nonstructural proteins: a 134–141 kDa protein terminated by an opal stop codon and a 194–201 kDa protein produced by readthrough of this stop codon, both of which are probably involved in RNA replication; a 29–30 kDa protein (P1a) “30K”-like cell-to-cell movement protein, which is involved in intercellular transport of the virus; and a 12–16 kDa protein (P1b), which is a suppressor of RNA silencing and determinant of seed transmission (of pea early-browning virus (PEBV) in pea). In addition to the virion structural protein, RNA-2 codes for two nonstructural proteins, P2b and P2c. The size of P2b ranges from 27 to 40 kDa in different isolates, and that of P2c from 18 to 33 kDa. P2b is absolutely required for transmission by nematodes, whereas mutation of the P2c gene affects nematode transmission in some strains but not in others. The genes for P2b and P2c are missing from some laboratory strains that have been maintained by mechanical transmission. RNA-2 of some tobravirus isolates contains an additional small ORF between the CP and P2b genes, which codes for a potential 9 kDa protein. RNA2 of tobacco rattle virus (TRV) isolate SYM has an unusual gene organization, with additional, novel genes being located 5′ of the CP gene.
Genome organization and replication
RNA-1 is capable of independent replication and systemic spread in plants. The 134–141 kDa and 194–201 kDa replication proteins are translated directly from it, whereas P1a and P1b are translated from sgRNA species 1a and 1b, respectively. RNA-2 does not itself have messenger activity; the CP is translated from sgRNA-2a. The means by which the other RNA-2 encoded proteins are expressed is not known but probably also involves sgRNAs (Figure 13). There is sequence homology between RNA-1 and RNA-2 at both ends, but the extent of the homology varies between strains. In some strains, the homologous region at the 3′ end is large enough to include some or all of the P1a and P1b genes of RNA-1, but it is not known if these genes are expressed from RNA-2. Accumulation of virus particles is sensitive to cycloheximide but not to chloramphenicol, suggesting that cytoplasmic ribosomes are involved in viral protein synthesis. Virions accumulate in the cytoplasm. L particles of pepper ringspot virus (PepRSV) become radially arranged around mitochondria, which are often distorted, and in cells infected with some TRV isolates, “X-bodies” largely composed of abnormal mitochondria and containing small aggregates of virus particles may be produced.
Antigenic properties
Tobravirus particles are moderately immunogenic. There is little or no serological relationship between members of the genus, and considerable antigenic heterogeneity among different isolates of the same virus.
Biological properties
The host ranges are wide, including members of more than 50 monocotyledonous and dicotyledonous plant families. The natural vectors are nematodes in the genera Trichodorus and Paratrichodorus (Trichodoridae), different species being specific for particular virus strains. Adults and juveniles can transmit, but virus is probably not retained through the molt. Ingested virus particles become attached to the esophageal wall of the nematodes, and are thought to be released by salivary gland secretions and introduced into susceptible root cells during exploratory feeding probes. Virus can be retained for many months by non-feeding nematodes. There is no evidence for multiplication of virus in the vector and it is probably not transmitted through nematode eggs. The viruses are transmitted through seed of many host species. TRV occurs in Europe (including Russia), Japan, New Zealand and North America; PEBV occurs in Europe and North Africa, and PepRSV occurs in South America. TRV causes diseases in a wide variety of crop plants as well as weeds and other wild plants, including spraing (corky ringspot) and stem mottle in potato, rattle in tobacco, streaky mottle in narcissus and tulip, ringspot in aster, notched leaf in gladiolus, malaria in hyacinth and yellow blotch in sugar beet. PEBV is the cause of diseases in several legumes, including broad bean yellow band, distorting mosaic of bean and pea early-browning. PepRSV causes diseases in artichoke, pepper and tomato.
Most tissues of systemically invaded plants can become infected, but in many species virus remains localized at the initial infection site in the roots. In some virus–host combinations, notably TRV in some potato cultivars, limited systemic invasion occurs, and virus may not be passed on to all the vegetative progeny of infected mother plants.
Normal particle-producing isolates (called M-type) are readily transmitted by inoculation with sap and by nematodes. Other isolates (called nm-type) have only RNA-1, do not produce particles, are transmitted with difficulty by inoculation with sap, and are probably not transmitted by nematodes. nm-type isolates are obtained from M-type isolates by using inocula containing only L particles, and are also found in naturally infected potato plants. They often cause more necrosis in plants than do their parent M-type cultures.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Nucleotide sequences of RNA-1 show <75% identity
- Interspecific pseudo-recombinant isolates cannot be made
- Host ranges differ in specific hosts (e.g. legumes)
- RNA-2 sequences and serological relationships are of limited value
List of species in the genus Tobravirus
| Pea early-browning virus | ||
| Pea early-browning virus-SP5 | [X14006=NC_002036 + X51828=NC_001368] | (PEBV-SP5) |
| Pepper ringspot virus | ||
| Pepper ringspot virus-CAM | [L23972=NC_003669 + X03241=NC_003670] | (PepRSV-CAM) |
| Tobacco rattle virus | ||
| Tobacco rattle virus-PpK20 | [AF166084=NC_003805 + Z36974=NC_003811] | (TRV-PpK20) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Tobravirus but have not been approved as species
None reported.
List of unassigned species in the family Virgaviridae
None.
List of other related viruses which may be members of the family Virgaviridae but have not been approved as species
| Nicotiana velutina mosaic virus | [D00906*] | (NVMV) |
* Sequence does not comprise the complete genome.
Phylogenetic relationships within the family
Best phylogenetic trees are obtained using the replication protein (or conserved domains within it) as these occupy the majority of the genome. However, the different genera usually separate reliably, regardless of the gene used (Figure 14). In both the replication protein and the coat protein, the monopartite genus Tobamovirus separates substantially from the remaining genera. Furovirus and Pomovirus occur together on the same branch in all trees as do Pecluvirus and Hordeivirus.
Within genera, only the genus Tobamovirus is large enough for particular subgroupings to be distinguished. Here, there are clearly groupings of closely-related viruses infecting similar hosts. The most obvious are those infecting cucurbits (CFMMV, CGMMV, KGMMV, ZGMMV), cruciferous plants (RMV, TVCV, WMoV, YoMV) and solanaceous plants (BrMMV, ObPV, PaMMV, PMMoV, TMGMV, TMV, ToMV).
Similarity with other taxa
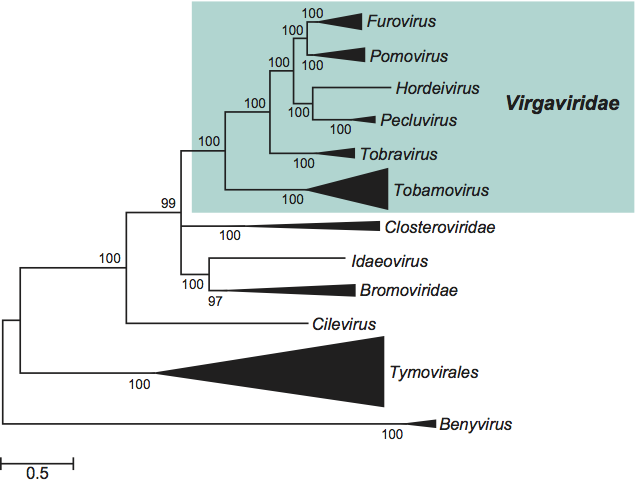
The replication proteins are related to those of other viruses with alpha-like replicases, and more particularly to the families Closteroviridae and Bromoviridae and the genus Idaeovirus. The only genus with rod-shaped virions excluded from the family is Benyvirus, because this is much more distantly related in phylogenetic analyses and because (unlike the members of the Virgaviridae) its members have a polyadenylated genome and a polymerase that is processed by autocatalytic protease activity (Figure 15).
Derivation of names
Furo: from fungus-borne, rod-shaped virus.
Hordei: from hordeus, Latin name of the primary host of the type species virus of the genus Hordeivirus.
Peclu: from peanut clump virus.
Pomo: from potato mop-top virus.
Tobra : tobacco rattle virus.
Tobamo: from tobacco mosaic virus.
Further reading
Adams, M.J., Antoniw, J.F., Kreuze, J., 2009. Virgaviridae: a new family of rod-shaped plant viruses. Arch. Virol. 154, 1967–1972.
Gibbs, A., 1999. Evolution and origins of tobamoviruses. Phil. Trans. R. Soc. Lond. B. 354, 593–602.
Jackson, A.O., Lim, H.-S., Bragg, J., Ganesan, U., Lee, M.Y., 2009. Hordeivirus replication, movement and pathogenesis. Annu. Rev. Phytopathol. 47, 385–422.
MacFarlane, S.A., 1999. Molecular biology of the tobraviruses. J. Gen. Virol. 80, 2799–2807.
Torrance, L., Lukhovitskaya, N.I., Schepetilnikov, M.V., Cowan, G.H., Ziegler, A., Savenkov, E.I., 2009. Unusual long-distance movement strategies of Potato mop top virus RNAs in Nicotiana benthamiana. MPMI. 22, 381–390.
Contributed by
Adams, M.J., Heinze, C., Jackson, A.O., Kreuze, J.F., Macfarlane, S.A. and Torrance, L.
Figures
Figure 1 Negative contrast electron micrograph of stained (ammonium molybdate pH 7.0) particles of soil-borne wheat mosaic virus (SBWMV). The bar represents 200 nm. Inset: Negative contrast electron micrograph of particles SBWMV stained with 1% uranyl acetate. The bar represents 100 nm.
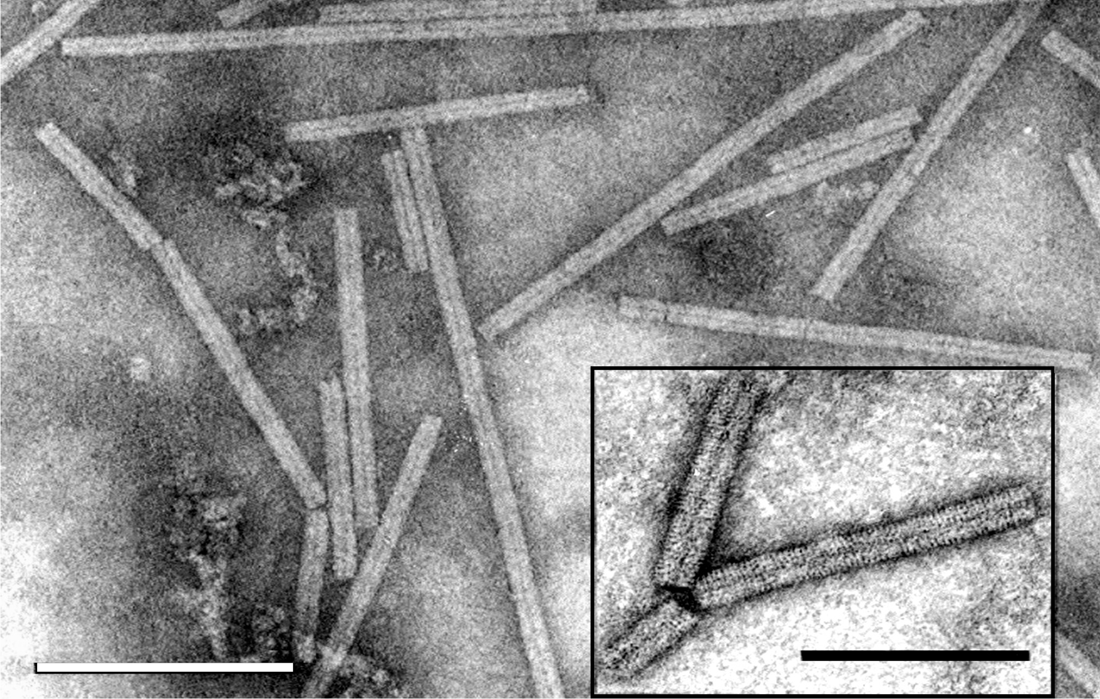
Figure 2 Genome organization of soil-borne wheat mosaic virus (SBWMV). The tRNA structure motifs at the 3-ends of the RNAs are represented by a dark square, the methyl transferase (Met), Helicase (Hel) and RNA-dependent RNA polymerase (RdRp) motifs by asterisks and the readthrough of the polymerase and coat protein ORFs by RT and an arrow.

Figure 3 Percentage sequence identities of total RNAs of furoviruses.

Figure 4 Electron micrograph of purified barley stripe mosaic virus (BSMV) particles stained with 2% uranyl acetate. The particles are approximately 20 nm wide and have a length that varies depending on the size of the encapsidated RNA. The field was selected to represent monomers, but often a range of heterodisperse end-to-end aggregates up to 1000 nm in length predominate in purified preparations. The particles in the top left, bottom center, and upper left side of the micrograph are end-to-end aggregates that occur during purification. The bar represents 150 nm.

Figure 5 Genome organization of barley stripe mosaic virus (BSMV). The color rectangles and smaller solid black rectangles represent the ORFs, and the 3-terminal tRNA-like structure, respectively. The 3-proximal ORFs on each RNA terminate with an UAA that initiates the short poly (A) tract that directly precedes the 238 nt tRNA-like terminus. RNA encodes a single protein, a, with an amino-terminal methyl transferase domain (Mtr) and a carboxy-terminal helicase domain (Hel). This protein is referred to as the helicase subunit of the replicase (RdRp). RNA encodes five proteins: a, the CP is translated from the genomic RNA; b, a 58 kDa protein that contains a helicase domain. b is translated from sgRNA1, whose promoter resides between positions 29 to 2 relative to the transcription start site; c, a 17 kDa protein that is separated from the b ORF by 173 nt; d, a 14 kDa protein which overlaps the b and the c ORFs; and d, a 23 kDa translational extension product of unknown function. The d, d, and c proteins are translated from sgRNA2. The sgRNA2 promoter is located between nt 32 to 17 relative to its transcription start site. RNA encodes two proteins. The a protein contains the GDD domain and is the polymerase subunit of the replicase. The cysteine-rich, 17 kDa b protein has RNA binding ability, and is translated from a sgRNA, whose promoter lies between positions 21 to +2 relative to the transcription start site.
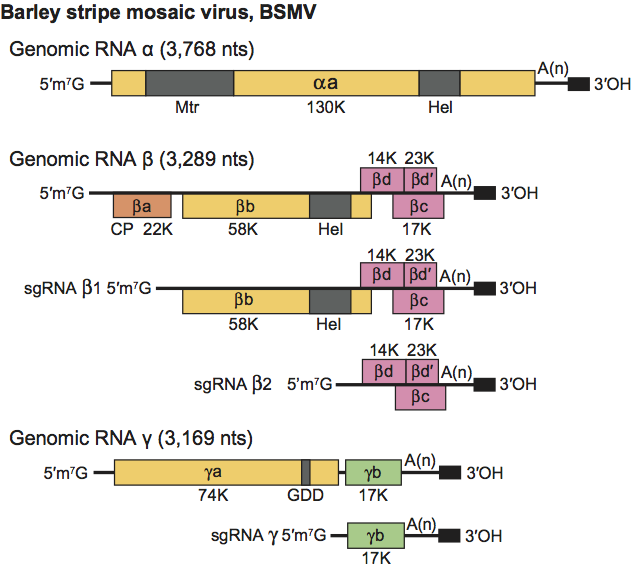
Figure 6 Negative contrast electron micrograph of virions of Indian peanut clump virus (L serotype) negatively stained with 2% phosphotungstic acid, pH 6. The bar represents 150 nm.
(Courtesy, G.H. Duncan.)

Figure 7 Genomic organization of peanut clump virus (PCV) RNAs. ORFs are indicated by rectangles and suppressible termination codon by an arrow (RT=readthrough).

Figure 8 Negative contrast electron micrograph of particles of potato mop-top virus. The gold-labeling shows the binding of monoclonal antibody SCR 68 to one extremity of the particles. The bar represents 100 nm.
(Courtesy I.M. Roberts.)
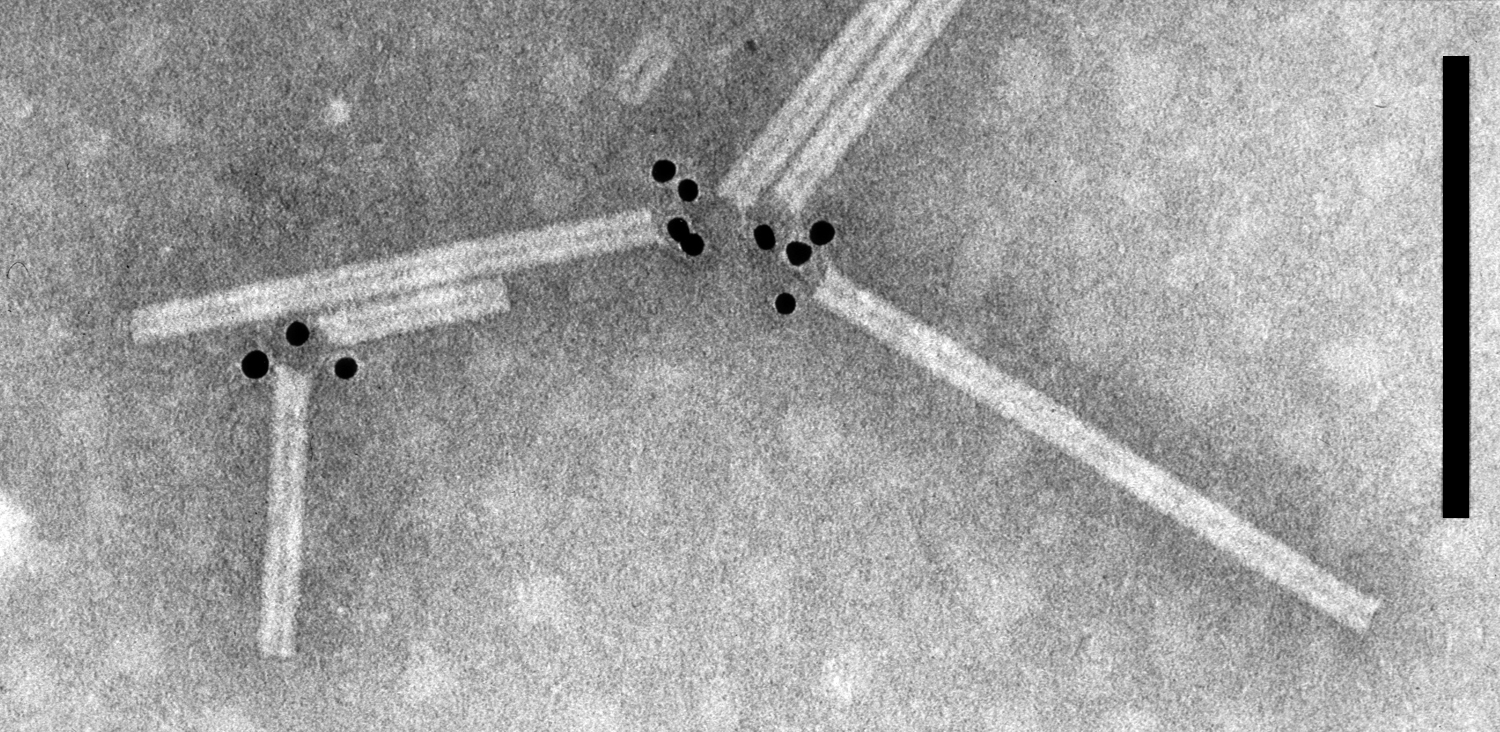
Figure 9 Genome organization typical of potato mop-top virus. Arrows indicate respectively the UGA and UAG stop codons that are thought to be suppressible, and solid squares indicate a 3-terminal tRNA-like structure. Hel, helicase; Mt, methyltransferase; RdRp, RNA dependent RNA polymerase; RT, readthrough.
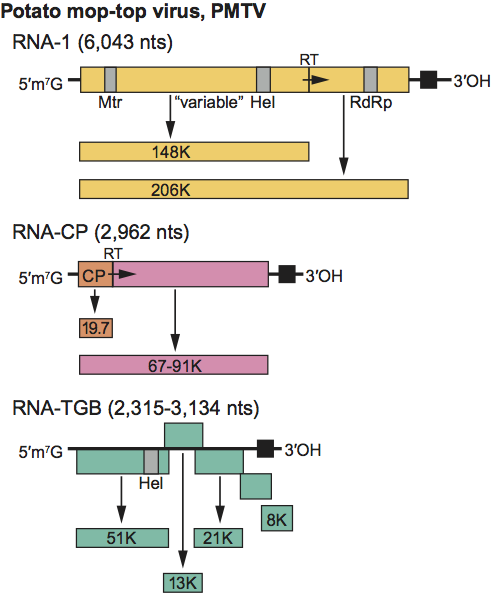
Figure 10 (Left) Model of particle of tobacco mosaic virus (TMV). Also shown is the RNA as it is thought to participate in the assembly process. (Right) Negative contrast electron micrograph of TMV particle stained with uranyl acetate. The bar represents 100 nm.

Figure 11 Genome organization of tobacco mosaic virus (TMV). Conserved replicase domains are indicated as shaded boxes. Genomic RNA is capped and is template for expression of the 126 and 183 kDa proteins. The 3 distal movement and CP ORFs are expressed from individual 3 co-terminal sgRNAs. CP=coat protein; MP=movement protein.
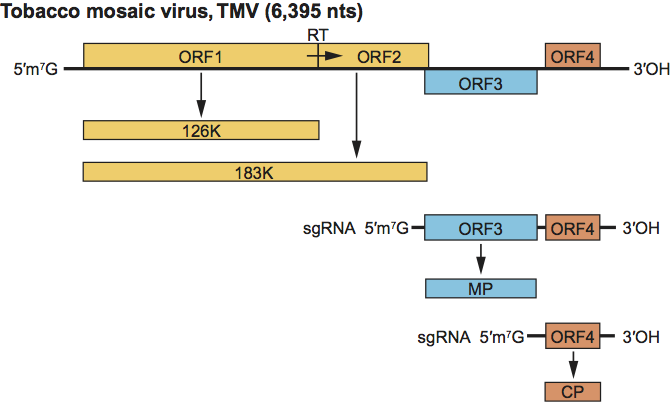
Figure 12 (Left) Diagram of a virion of tobacco rattle virus (TRV), in section. (Right) Negative contrast electron micrograph of particles of TRV. The bar represents 100 nm.

Figure 13 Genome organization and strategy of expression of tobacco rattle virus (TRV). The means by which P2b and P2c are expressed is unknown.
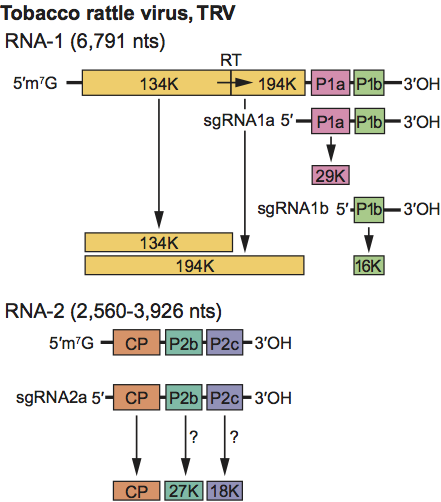
Figure 14 Phylogenetic (distance) trees based on the amino acid sequences of the entire replication protein, the Triple Gene Block protein 1 (TGB1), the movement protein and the coat protein. A single representative isolate of each sequenced species in the family was included. Numbers on branches indicate percentage of bootstrap support out of 1,000 bootstrap replications (when >60%). The scale indicates JTT amino acid distances. Trees produced in MEGA4. The BSMV replication protein was combined from two different genome components.
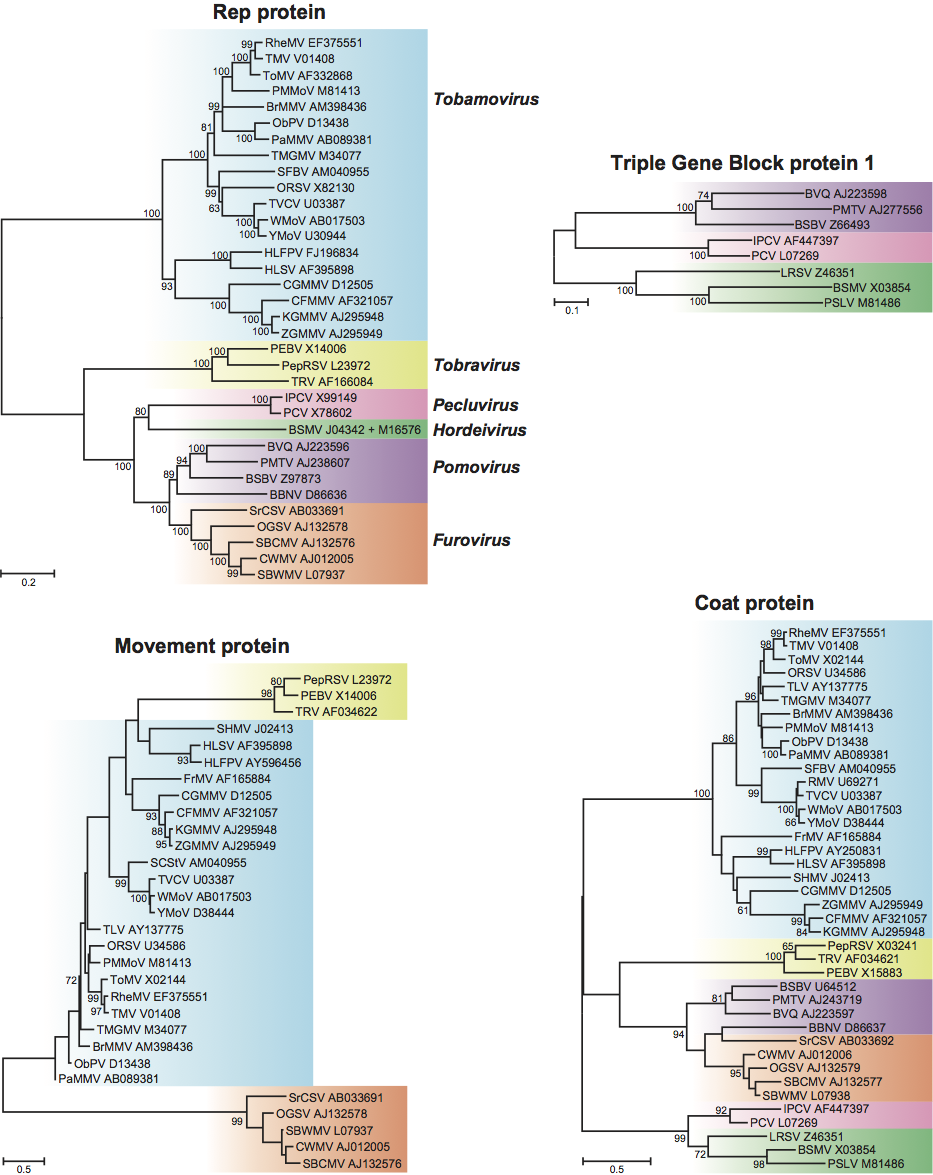
Figure 15 Bayesian phylogenetic tree of the nucleotide sequences of the fused MetHelRdRp domains of the members of the six genera included in the family Virgaviridae together with some other related viruses. A total of 500 amino acid positions corresponding to 1,500 nt positions were used for the alignment. The tree was generated from a back-translated amino-acid alignment using MrBayes v3.1.2, employing the general time reversible model with gamma-shaped rate variation with a proportion of invariable sites; 1,000,000 generations of MCMC analysis were performed, to the point at which the average standard generation of split frequency between two parallel runs had reached 0.009565. Posterior probability values are indicated on the corresponding branches. Nearly identical trees were obtained by neighbour-joining and maximum composite likelihood methods. Genera and families (which are all monophyletic) have been collapsed into a triangle, the length of which corresponds to the variation found within the clade.