Genus: Umbravirus
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Type species: Carrot mottle virus
Distinguishing features
Positive sense, single strand RNA viruses which do not form conventional virus particles and lack a coat protein ORF but are encapsidated with a helper virus.
Virion properties
Morphology
Umbraviruses do not form conventional virus particles, and the five genomes whose complete sequences are known lack plausible ORFs for capsid CPs. Umbraviruses rely on the CP of a helper virus, characteristically from a virus in the family Luteoviridae, for encapsidation and for transmission by the vector of the helper virus. However, in single infections by umbraviruses, the infectivity in buffer extracts of leaves is surprisingly stable, though very sensitive to treatment with organic solvents, suggesting that the infective RNA is protected in lipid-containing structures. In plants infected with carrot mottle virus (CMoV), enveloped structures about 52 nm in diameter (Figure 1A, B) occur in the vacuoles of infected cells and in partially purified preparations. These structures may be involved in virus replication and/or serve to protect the RNA. Similar structures occur in plants infected with the bean yellow vein-banding strain of Pea enation mosaic virus-2 (BYVBV), groundnut rosette virus (GRV) and lettuce speckles mottle virus (LSMV), but no information is available for other umbraviruses. An infective fraction from GRV-infected tissues contained filamentous ribonucleoprotein (RNP) particles (Figure 1C) composed of viral RNA, the umbraviral ORF3 protein and the host nucleolar protein, fibrillarin.
Physicochemical and physical properties
Infectivity in leaf extracts is stable for several hours at room temperature or several days at 5 °C, but is abolished by treatment with organic solvents. Partially purified preparations of CMoV consist predominantly of cell membranes but contain infective components which, because they have a sedimentation coefficient of about 270S and a buoyant density of about 1.15 g cm−3 in CsCl, are probably the 52 nm-diameter enveloped structures observed in these preparations (Figure 1A, B). An infective fraction from GRV-infected tissue contained complexes with a buoyant density of 1.34–1.45 g cm−3 consisting of filamentous RNP particles, composed of the umbraviral ORF3 protein, host fibrillarin and virus RNA (Figure 1C). The relationship between the enveloped and filamentous structures is unclear.
Nucleic acid
Nucleic acid preparations made by extracting leaves with phenol are often much more infective than buffer extracts. The infective agent in these preparations is a ssRNA, but the preparations also contain abundant dsRNA. The genome consists of one linear segment of positive-sense ssRNA 4.0–4.2 kb in length. Umbravirus genomic RNAs are not polyadenylated at their 3′ ends and lack a 5′ cap structure. The 3′ UTR of pea enation mosaic virus (PEMV-2) genomic RNA contains the 100 nt cap-independent translational element which binds the eIF4E subunit of eukaryotic translation initiation factor eIF4F. In the case of GRV, a 900 nt ssRNA satellite RNA is essential for the encapsidation of the GRV genomic RNA with the coat protein of groundnut rosette assistor virus (unassigned member of the family Luteoviridae).
Proteins
No conventional structural proteins are reported. The nucleotide sequences lack plausible ORFs for CPs but possess ORFs for four potential non-structural protein products (Figure 2).
Lipids
Although no conventional virus particles are formed, the sensitivity to organic solvents, and low buoyant density, of the infective components in partially purified preparations of CMoV suggests that this infectivity is associated with lipid, probably of plant origin. The infective components probably correspond to the enveloped structures seen in sections of infected leaves.
Carbohydrates
None reported.
Genome organization and replication
Figure 2 shows the genome organization of GRV; those of other umbraviruses are very similar. For each RNA, there is at the 5′ end a very short non-coding region preceding ORF1, which encodes a putative product of 31–37 kDa. ORF2, which slightly overlaps the end of ORF1, could encode a product of 63–65 kDa but lacks an AUG initiation codon near its 5′ end. However, immediately before the stop codon of ORF1 there is a 7 nt sequence that is associated with frameshifting in several plant and animal viruses, and it seems probable that ORF1 and ORF2 are translated as a single polypeptide of 94–98 kDa by a mechanism involving a −1 frameshift. The predicted product contains, in the ORF2 region, sequence motifs characteristic of viral RdRp. A short untranslated region separates ORF2 from ORF3 and ORF4, which overlap each other almost completely in different reading frames and each yield a putative product of 26–29 kDa. The ORF4 product contains sequences characteristic of plant virus MPs. The ORF3 product of different umbraviruses studied have up to 50% similarity to each other but no significant similarity to any other viral or non-viral proteins; their function is to protect viral RNA and enable its transport through the phloem.
Umbravirus-infected leaf tissue contains abundant dsRNA that is not itself infective but that becomes so when heat-denatured. Two dsRNA species are common to all umbraviruses: dsRNA-1 (ca. 4.2–4.8 kbp) and dsRNA-2 (ca. 1.1–1.5 kbp). cDNA copies of dsRNA-1 hybridize with dsRNA-2 and these molecules are thought to represent double stranded forms of, respectively, genomic and subgenomic ssRNA species. ORFs 3 and 4 are probably expressed from sgRNA. There is evidence for the presence in GRV-infected plants of two less-than-full-length RNA species of very similar size, close to that expected for such sgRNAs, and corresponding to that of dsRNA-2. The dsRNA-2 of CMoMV has been shown to include the sequences of ORFs 3 and 4, and the 3′ UTR.
Some umbraviruses possess one or more additional dsRNA species, associated in at least one instance (GRV) with the presence of a satellite RNA. PEMV-2 also has a satellite RNA, and each of these satellites can be supported by the helper virus of the other.
Antigenic properties
None reported.
Biological properties
Host range
Individual umbraviruses are confined in nature to one or a few host plant species. Their experimental host ranges are broader but still restricted. The symptoms induced in infected plants are usually mottles or mosaics. Symptoms of GRV are greatly influenced by the associated satellite RNA.
Transmission
Umbraviruses are transmissible, sometimes with difficulty, by mechanical inoculation. However, in nature each is dependent on a specific helper virus for transmission in a persistent (circulative, non-propagative) manner by aphids. All helper viruses that have been characterized are members of the family Luteoviridae. The mechanism of this dependence is encapsidation of the dependent virus RNA in the CP of the helper. In GRV, the satellite RNA plays an essential role in mediating this luteovirus-dependent aphid transmission. There is no evidence for multiplication of umbraviruses in the insect vector. Seed transmission has not been reported.
Geographical distribution
CMoV and/or carrot mottle mimic virus and PEMV-2 apparently occur worldwide wherever their crop hosts are grown; other umbraviruses have a restricted distribution. Several umbraviruses, notably GRV, occur only in Africa.
Pathogenicity
Although all umbraviruses depend on a helper virus, a member of the family Luteoviridae, for transmission by vector insects, several of them are as important as, or more important than, their helpers in the causation of disease symptoms. The umbravirus of greatest economic importance is GRV, which causes the most devastating virus disease of groundnut (peanut) in Africa. However, in this case it is a GRV-dependent satellite RNA that is the actual cause of the symptoms. In most instances umbraviruses have not been shown to contribute functions essential for the biological success of their associated helper viruses. However, a notable exception is PEMV-2, which is essential for the systemic spread of PEMV-1 in plants. PEMV-2 even allows PEMV-1 and another member of the family Luteoviridae, potato leafroll virus, to spread out of the phloem into mesophyll tissue and thereby to become transmissible by manual inoculation. Similarly, another member of the Luteoviridae, beet western yellows virus, has been reported to show limited manual transmissibility when in the presence of the umbravirus LSMV.
Cytopathic effects
Umbraviruses, even in the absence of their helper viruses, exhibit rapid systemic spread in plants. They infect cells throughout the leaf, though presumably the aphid transmissible particles are restricted to the same tissues (in most instances the phloem) as the luteoviruses that provide their CP. In mesophyll cells infected with CMoV there is extensive development of cell wall outgrowths sheathing elongated plasmodesmatal tubules. The ORF3 protein of GRV targets and reorganizes Cajal bodies (CB) into multiple CB-like structures and then enters the nucleolus by causing fusion of these structures with the nucleolus. The nucleolar localization of the ORF3 protein is essential for subsequent formation of viral RNP particles capable of virus long-distance movement and systemic infection.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Natural host range
- dsRNA band pattern (bearing in mind that some bands may represent satellite RNA species)
- Nucleotide sequence identity less than 70%
- Little or no hybridization with cDNA probes representing most parts of the genome
List of species in the genus Umbravirus
| Carrot mottle mimic virus |
|
|
| Carrot mottle mimic virus-Australia | [U57305=NC_001726] | (CMoMV-AU) |
| Carrot mottle virus |
|
|
| Carrot mottle virus-Weddel | [FJ188473=NC_011515] | (CMoV-We) |
| Groundnut rosette virus |
|
|
| Groundnut rosette virus-MC1 | [Z69910= NC_003603] | (GRV-MC1) |
| Lettuce speckles mottle virus |
|
|
| Lettuce speckles mottle virus-California |
| (LSMV-CA) |
| Pea enation mosaic virus-2 |
|
|
| Bean yellow vein-banding virus |
| (BYVBV) |
| Pea enation mosaic virus-2-WGS | [U03563] | (PEMV-2-WGS) |
| Tobacco bushy top virus |
|
|
| Tobacco bushy top virus-Baoshan | [AF431890= NC_004366] | (TBTV-Bao) |
| Tobacco mottle virus |
|
|
| Tobacco mottle virus-Zimbabwe | [AY007231*] | (TMoV-ZW) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequence does not comprise the complete genome.
List of other related viruses which may be members of the genus Umbravirus but have not been approved as species
| Opium poppies mosaic virus | [EU151723*] | (OPMV) |
| Sunflower crinkle virus |
| (SuCV) |
| Sunflower yellow blotch virus |
| (SuYBV) |
| Tobacco yellow vein virus |
| (TYVV) |
* Sequence does not comprise the complete genome.
Similarity with other taxa
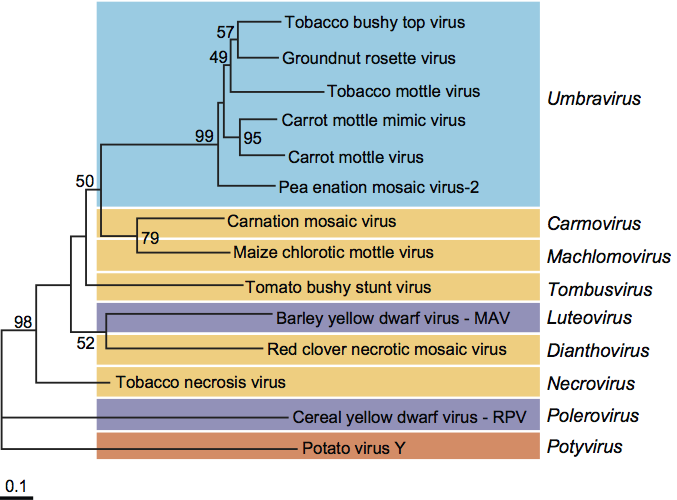
Amino acid sequence comparisons show that the putative RdRps encoded by the genomic RNA of umbraviruses belong to the so-called supergroup 2 of RNA polymerases, as do those of viruses in the genera Carmovirus, Dianthovirus, Luteovirus, Machlomovirus, Necrovirus and Tombusvirus (Figure 3). Since these enzymes are the only universally conserved proteins of positive strand RNA viruses, the genus Umbravirus might be considered to be in or close to the family Tombusviridae.
Derivation of names
Umbra: from Latin umbra, “a shadow”. In English, a shadow is an uninvited guest that comes with an invited one.
Further reading
Adams and Hull, 1972 A.N. Adams, R. Hull, Tobacco yellow vein, a virus dependent on assistor viruses for its transmission by aphids. Ann. Appl. Biol. 71 (1972) 135–140.
Demler et al., 1996 S.A. Demler, G.A. de Zoeten, G. Adam, K.F. Harris, Pea enation mosaic enamovirus: properties and aphid transmission. In: . Plenum Press, New York1996303–344.
Gibbs et al., 1996 M.J. Gibbs, J.I. Cooper, P.M. Waterhouse, The genome organization and affinities of an Australian isolate of carrot mottle umbravirus. Virology. 224 (1996) 310–313.
Kim et al., 2007 S.H. Kim, E.V. Ryabov, N.O. Kalinina, D.V. Rakitina, T. Gillespie, S. MacFarlane, S. Haupt, J.W. Brown, M. Taliansky, Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 26 (2007) 2169–2179.
Murant, 1993 A.F. Murant, R.E.F. Matthews, Complexes of transmission-dependent and helper virusesDiagnosis of Plant Virus Diseases. In: R.E.F. Matthews, Diagnosis of Plant Virus Diseases. CRC Press, Boca Raton, FL1993333–357.
Murant et al., 1969 A.F. Murant, R.A. Goold, I.M. Roberts, J. Cathro, Carrot mottle – a persistent aphid-borne virus with unusual properties and particles. J. Gen. Virol. 4 (1969) 329–341.
Murant et al., 1988 A.F. Murant, R. Rajeshwari, D.J. Robinson, J.H. Raschké, A satellite RNA of groundnut rosette virus that is largely responsible for symptoms of groundnut rosette disease. J. Gen. Virol. 69 (1988) 1479–1486.
Smith, 1946 K.M. Smith, The transmission of a plant virus complex by aphides. Parasitology. 37 (1946) 131–134.
Taliansky et al., 2003 M.E. Taliansky, I.M. Roberts, N.O. Kalinina, E.V. Ryabov, S.K. Raj, D.J. Robinson, K.J. Oparka, An umbraviral protein, involved in long-distance RNA movement, binds viral RNA and forms unique, protective ribonucleoprotein complexes. J. Virol. 77 (2003) 3031–3040.
Wang et al., 2009 Z. Wang, K. Treder, W.A. Miller, Structure of a viral cap-independent translation element that functions via high affinity binding to the eIF4E subunit of eIF4F. J. Biol. Chem. 284 (2009) 14189–14202.
Contributed by
Ryabov, E.V., Taliansky, M.E., Robinson, D.J., Waterhouse, P.M., Murant, A.F., de Zoeten, G.A., Falk, B.W., Vetten, H.J. and Gibbs, M.J.
Figures
Figure 1 (A) Section of palisade mesophyll cell from a leaf of Nicotiana clevelandii systemically infected with carrot mottle virus (CMoV), showing enveloped structures (E) about 52 nm in diameter in the cell vacuole (V) in association with the tonoplast (T). The bar represents 250 nm. (B) Enveloped structures about 52 nm in diameter in a partially purified preparation from CMoV-infected N. clevelandii, stained with 2% uranyl acetate. The bar represents 100 nm. (C) Section of a Nicotiana benthamiana cell expressing the GRV ORF3 protein. The section shows filamentous ribonucleoprotein particles embedded in an electron-dense matrix. The section was labeled by in situ hybridization with a GRV-specific RNA probe. The bar represents 100 nm.
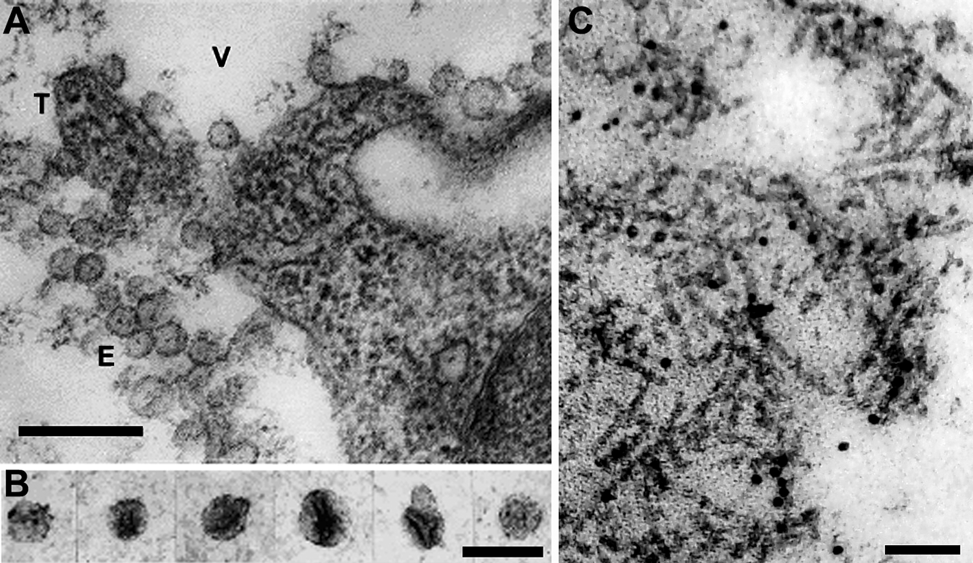
Figure 2 Diagram showing the genomic organization of groundnut rosette virus (GRV). The continuous horizontal line represents the genome RNA, and the numbered blocks the correspondingly numbered ORFs. The lower part of the diagram shows the predicted translation products, with their size. The potential product of ORF1 has not been shown to exist. The single product produced from ORFs 1 and 2, probably as a result of a 1 frameshift event (FS), is thought to be a polymerase because it contains, in the ORF2 region, sequences characteristic of viral RdRp. The ORF3 product functions to protect viral RNA and enable its transport through the phloem. The ORF4 product, marked MP, has a cell-to-cell movement function.
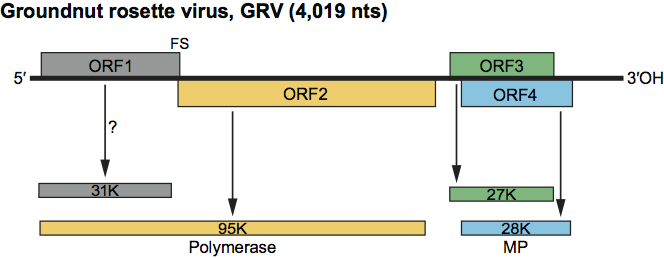
Figure 3 Phylogenetic relationships of the RdRps of umbraviruses and some other plant viruses. Amino acid sequences were aligned using the ClustalX2 program. The neighbour-joining trees were produced and bootstrapped using the PHYLIP package programs. Numbers at the nodes represent bootstrap values as percentages obtained from 2000 replications, shown only for branches supported by more than 40%. Length of branches is proportional to number of changes.