Family: Tymoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Distinguishing features
This is the only family within the order Tymovirales that has viruses with isometric particles. The replication protein polymerase domain forms a distinctive phylogenetic lineage.
Virion properties
Morphology
Virions are isometric, non-enveloped, about 30 nm in diameter, with a rounded contour and prominent surface structure, with clustering of CP subunits in pentamers and hexamers. The capsids of tymoviruses are made up of 20 hexameric and 12 pentameric subunits arranged in a T=3 icosahedron and the RNA appears to be at least partially ordered in an icosahedral arrangement in the center of the protein shell. (See Figure 1.)
Physicochemical and physical properties
Virus particles sediment as two centrifugal components: T, made up of non-infectious protein shells that contain little or no RNA (primarily subgenomic CP mRNA), and B, composed of intact nucleoprotein particles. Sedimentation coefficients (Sw,20) of component T and B range from 42 to 55 and from 109 to 125, respectively. Buoyant densities in CsCl of components T and B are 1.26–1.28 g cm−3 and 1.40–1.46 g cm−3 respectively. Virions resist high temperatures (thermal inactivation point is 60–65 °C up to above 80 °C with some tymoviruses) and organic solvents, but are disrupted by SDS.
Nucleic acid
Virions contain a single molecule of positive sense, ssRNA constituting 25–35% of the particle weight. The RNA has a very high cytidine content (from 32 to about 50%) and ranges from 6.0 to 7.5 kb in length. Tymovirus genomes are capped at the 5′ terminus with m7G and most, though not all, have a tRNA-like structure at the 3′ end, which for turnip yellow mosaic virus (TYMV) and several other members, accepts valine. The genomes of marafiviruses, maculaviruses and of the unassigned Poinsettia mosaic virus (PnMV) are polyadenylated at the 3′ terminus and are thought to be capped at the 5′ end with m7G.
Proteins
The CP of virus particles consists of either a single protein species with molecular mass of 20 kDa (tymoviruses), 24.5–25 kDa (maculaviruses), or a major protein of about 21 kDa and a minor protein of 22.5–25 kDa (marafiviruses). The capsids of PnMV and a strain of Bermuda grass etched line virus are made up of a single protein of about 21 kDa.
Genome organization and replication
The genomes of tymoviruses contain two extensively overlapping ORFs that begin 7 nt apart, and a third ORF (for CP) that is expressed from a 3′-coterminal subgenomic RNA (sgRNA). The longest ORF encodes a replication polyprotein of about 220 kDa that contains recognized domains for a methyltransferase (Mtr), a papain-like cysteine protease (P-Pro), an RNA helicase (Hel) and an RNA-dependent RNA polymerase (RdRp). The genomes of the marafiviruses, maculaviruses and PnMV possess a similar polyprotein ORF, and a CP ORF in the same reading frame that is either fused to the long ORF (marafiviruses and PnMV) or separated from it by two adjacent stop codons (grapevine fleck virus, GFkV). MRFV is the sole marafivirus with an extensive ORF near the 5′ end of the genome that entirely overlaps the replication protein ORF (see Figure 3 below). The genome of GFkV possesses two additional ORFs towards the 3′ end. Viral RNA of tymoviruses replicates in the cytoplasm in association with the double-membraned invaginations that line the periphery of the chloroplasts. Comparable vesicles occurring at the peripheries of mitochondria or chloroplasts of cells infected with maculaviruses and PnMV may have the same function. Genome expression of the tymoviruses includes post-translational autocatalytic cleavage of the replication polyprotein by the P-Pro, and synthesis and translation of a 3′-coterminal sgRNA for CP expression.
Antigenic properties
Virions are moderately to highly antigenic. Tymoviruses cluster roughly into two serological groups with cross-reactivity ranging from strong to weak within each group. Cross-reactivities between group members are weak to undetectable. Serological relationships occur between some marafiviruses, but not between individual maculaviruses.
Biological properties
Host range
Members of this family infect dicotyledonous plants, with the exception of some marafiviruses that primarily infect plants in the Gramineae. Natural and experimental host ranges of individual virus species are narrow, sometimes restricted to a single type of host (e.g., GFkV infects only Vitis; MRFV only Zea). Disease symptoms are bright yellow mosaic or mottling (tymoviruses and PnMV), chlorotic stripes, vein clearing, etched lines or dwarfing (marafiviruses), and flecking of the leaves (maculaviruses).
Transmission
Tymoviruses and PnMV, but not other members of the family, are readily transmissible by mechanical inoculation. They replicate to high titres and invade all main tissues of the host. Marafi- and maculaviruses are phloem-limited. Some marafiviruses (MRFV, oat blue dwarf virus (OBDV) and Bermuda grass etched-line virus (BELV)) are distinguished by being vectored in a persistent manner by leafhoppers, in which they replicate. Some tymoviruses are weakly seed-transmissible, and are also spread by beetles, which serve as low-efficiency local vectors. Maculaviruses do not have recognized vectors, and the grapevine-associated marafi- and maculaviruses are disseminated primarily through infected propagating material.
Geographical distribution
Members of the family have been recorded from most parts of the world. Geographical distribution of individual species varies from restricted to widespread.
Cytopathic effects
Most species elicit derangement of the internal structure and alteration of the shape of chloroplasts and/or mitochondria, which also show rows of peripheral vesicles derived from localized invaginations of the limiting membrane.
Genus demarcation criteria in the family
The criteria demarcating genera in the family are:
- Biological criteria: Tymoviruses are mechanically transmissible, invade parenchyma tissues, and infect dicotyledonous plants. Marafiviruses and maculaviruses are both phloem-limited and are not mechanically transmissible.
- Epidemiological criteria: Tymoviruses are transmitted by beetles, some marafiviruses by leafhoppers. Maculaviruses have no known vector.
- Cytopathological criteria: Tymoviruses elicit the formation of double-membraned vesicles at the periphery of chloroplasts while maculaviruses elicit them at the periphery of mitochondria. Chloroplast vesicles induced by PnMV are single-membraned.
- Physicochemical criteria: (a) Genome size: 6.0–6.7 kb (tymoviruses), 6.3–6.5 kb (marafiviruses), 7.5 kb (maculaviruses); (b) number and molecular mass of the CP: one subunit type of 20 kDa (tymoviruses), 25 kDa (maculaviruses) or 21 kDa (PnMV), or two subunit types (21 and 23–25 kDa) (marafiviruses).
- Molecular criteria: Tymovirus genomes have three ORFs and usually a tRNA-like structure at the 3′ terminus. Maculavirus genomes have four ORFs, a 3′-terminal poly(A) tail. Marafivirus genomes have a single large ORF (two ORFs in the case of MRFV) and a 3′-terminal poly(A) tail. Tymovirus genomes possess a conserved sequence that serves as a sgRNA promoter known as the “tymobox”. The genomes from marafiviruses and PnMV contain a variant termed the “marafibox”, while no such sequence is evident in maculavirus genomes.
Genus Tymovirus
Type species Turnip yellow mosaic virus
Distinguishing features
The genomic RNA (6.0–6.7 kb in size) contains three ORFs (Figure 2). ORF1 encodes the large replication polyprotein. The 3′ ends of these large ORFs are highly conserved and contain a 16 nt sequence known as the “tymobox” (GAGUCUGAAUUGCUUC), which functions as subgenomic RNA promoter. ORF2 initiates 7 nt upstream of ORF1, almost entirely overlaps ORF1, and encodes a 50–80 kDa proline-rich protein that for TYMV is dispensable for replication but is required for cell-to-cell movement. ORF3 codes for the viral CP (20 kDa), which is expressed via a sgRNA. The narrow host range of tymoviruses makes host susceptibility an important property in distinguishing among species. All members induce vesicles at the periphery of chloroplasts and to a lesser degree at mitochondria, resulting in characteristic aggregates of swollen and modified chloroplasts. All main tissues of the hosts are invaded. Empty virion shells sometimes accumulate in the nuclei. Beetles of the families Chrysomelidae and Curculionidae serve as local vectors, transmitting the virus in a semi-persistent manner. All members of the genus are mechanically transmissible and a few (TYMV, eggplant mosaic virus and Dulcamara mottle virus) are transmitted through seeds.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Overall genome sequence identity of less than 80%.
- Capsid protein sequences less than 90% identical.
- Differential host range.
- Differences in the 3′-terminal structure.
- Serological specificity.
List of species in the genus Tymovirus
| Anagyris vein yellowing virus |
|
|
| Anagyris vein yellowing virus - Apulia, Italy 1983 | [AY751780 = NC_011559] | (AVYV-IT) |
| Andean potato latent virus |
|
|
| Andean potato latent virus-Hu | [AF035402]* | (APLV-Hu) |
| Belladonna mottle virus |
|
|
| Belladonna mottle virus-Eur | [X54529]* | (BeMV-Eur) |
| Cacao yellow mosaic virus |
|
|
| Cacao yellow mosaic virus - West Africa | [X54354]* | (CYMV-WA) |
| Calopogonium yellow vein virus |
|
|
| Calopogonium yellow vein virus - Malaysia | [U91413]* | (CalYVV-MAL) |
| Chayote mosaic virus |
|
|
| Chayote mosaic virus - Costa Rica | [AF195000 = NC_002588] | (ChMV-CR) |
| Clitoria yellow vein virus |
|
|
| Clitoria yellow vein virus - Kenya | [AF035200]* | (CYVV-KE) |
| Desmodium yellow mottle virus |
|
|
| Desmodium yellow mottle virus - USA | [AF035201]* | (DYMoV-USA) |
| Dulcamara mottle virus |
|
|
| Dulcamara mottle virus - Hertfordshire, UK | [AY789137 = NC_007609] | (DuMV-UK) |
| Eggplant mosaic virus |
|
|
| Eggplant mosaic virus - Trinidad | [J04374 = NC_001480] | (EMV-TRD) |
| Erysimum latent virus |
|
|
| Erysimum latent virus - Eastern Germany | [AF098523 = NC_001977] | (ErLV-DE) |
| Kennedya yellow mosaic virus |
|
|
| Kennedya yellow mosaic virus-JB - Australia | [D00637 = NC_001746] | (KYMV-AUS) |
| Melon rugose mosaic virus |
|
|
| Melon rugose mosaic virus - Yemen |
| (MRMV-YE) |
| Nemesia ring necrosis virus |
|
|
| Nemesia ring necrosis virus - Bavaria, Germany | [AY751778 = NC_011538] | (NeRNV-DE) |
| Diascia yellow mottle virus DiaYMV-WA | [EU684141 = NC_011086] | (DiaYMV-WA) |
| Okra mosaic virus |
|
|
| Okra mosaic virus - Nigeria | [EF554577 = NC_009532] | (OkMV-NG) |
| Ononis yellow mosaic virus |
|
|
| Ononis yellow mosaic virus - England | [J04375 = NC_001513] | (OYMV-UK) |
| Passion fruit yellow mosaic virus |
|
|
| Passion fruit yellow mosaic virus - Colombia | [AF467107]* | (PFYMV-COL) |
| Peanut yellow mosaic virus |
|
|
| Peanut yellow mosaic virus |
| (PeYMV) |
| Petunia vein banding virus |
|
|
| Petunia vein banding virus - Rio Grande do Sol, Brazil | [AF210709]* | (PetVBV-BR) |
| Physalis mottle virus |
|
|
| Physalis mottle virus – Iowa (Belladonna mottle virus-Iowa) | [Y16104 = NC_003634] | (PhyMV-IO) |
| Plantago mottle virus |
|
|
| Plantago mottle virus - USA | [AY751779 = NC_011539] | (PlMoV-USA) |
| Scrophularia mottle virus |
|
|
| Scrophularia mottle virus - Germany | [AY751777 = NC_011537] | (SrMV-DE) |
| Turnip yellow mosaic virus |
|
|
| Turnip yellow mosaic virus - Europe | [X07441 = NC_004063] | (TYMV-Eur) |
| Turnip yellow mosaic virus-TYMC; infectious clone | [X16378] | (TYMV-C) |
| Turnip yellow mosaic virus-BL - Australia | [AF035403] | (TYMV-BL) |
| Voandzeia necrotic mosaic virus |
|
|
| Voandzeia necrotic mosaic virus - Côte d’Ivoire |
| (VNMV-CdI) |
| Wild cucumber mosaic virus |
|
|
| Wild cucumber mosaic virus - Oregon, USA | [AF035633]* | (WCMV-OR) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Tymovirus but have not been approved as species
| Chiltepin yellow mosaic virus | [FN563123 = NC_014127] | (ChiYMV) |
Genus Marafivirus
Type species Maize rayado fino virus
Distinguishing features
The distinct feature of the marafivirus genome (6.3–6.8 kb in size) is a large ORF encoding a precursor polypeptide consisting of the replication-associated proteins and the larger (23–25 kDa) of the two forms of CP found in the virions. The “marafibox” [CA(G/A)GGUGAAUUGCUUC] is a conserved 16 nt stretch comparable to the “tymobox”, from which it differs by three or four residue changes. The 23–25 kDa CP is produced initially as a C-terminal fusion of the replication protein, while the smaller (ca. 21 kDa) CP is produced from a 3′-co-terminal sgRNA. Viruses presently classified as marafiviruses exhibit some diversity in genome design (Figure 3): some marafivirus genomes possess a putative second ORF overlapping the main ORF either towards the 5′ end (MRFV) or 3′ end (citrus sudden death-associated virus). Host ranges are very narrow, making host susceptibility an important identifying criterion. Marafiviruses are strictly confined to the phloem of infected hosts and are not readily transmissible by sap inoculation, although MRFV and OBDV have been transmitted by vascular puncture. For MRFV, OBDV and BELV, transmission is by leafhoppers in a persistent-propagative manner, with virus replication occurring in the insect host. Species-specific virus-vector associations occur: i.e., MRFV is transmitted by Dalbulus, OBDV by Macrosteles, and BELV by Aconurella. The insect-transmitted species infect primarily plants in the Gramineae, although a notable exception is the infection of flax by OBDV. None of the species is transmitted through seeds. No insect vectors have been identified for other marafiviruses.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Overall genome sequence identity of less than 80%.
- Capsid protein sequences less than 90% identical.
- Differences in the 3′-terminal structure and in the number of ORFs.
- Differential host range.
- Vector specificity.
- Serological specificity.
List of species in the genus Marafivirus
| Bermuda grass etched-line virus |
|
|
| Bermuda grass etched-line virus - Morocco | [AY040531]* | (BELV-MOR) |
| Citrus sudden death-associated virus |
|
|
| Citrus sudden death-associated virus - Brazil | [AY884005 = NC_006950] | (CSDaV-BR) |
| Maize rayado fino virus |
|
|
| Maize rayado fino virus - Costa Rica | [AF265566 = NC_002786] | (MRFV-CR) |
| Oat blue dwarf virus |
|
|
| Oat blue dwarf virus - North Dakota, USA | [U87832 = NC_001793] | (OBDV-ND) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequence does not comprise the complete genome.
List of other related viruses which may be members of the genus Marafivirus but have not been approved as species
| Blackberry virus S | [FJ915122] | (BlVS) |
| Grapevine asteroid mosaic-associated virus | [AJ249357, AJ249358]* | (GAMaV) |
| Grapevine rupestris vein feathering virus | [AY706994] | (GRVFV) |
| Grapevine Syrah virus-1(Grapevine virus Q) | [FJ436028][FJ977041] | (GSyV-1) |
* Sequences do not comprise the complete genome.
Genus Maculavirus
Type species Grapevine fleck virus
Distinguishing features
The genomic RNA of GFkV (7.5 kb in size) is the largest in the family, contains four ORFs and is 3′-polyadenylated (Figure 4). ORF1 encodes the replication polyprotein but does not seem to have a conserved sequence comparable to the “tymobox” or the “marafibox”. ORF2 codes for the 24 kDa CP. ORF3 and ORF4, which are located at the extreme 3′ end of the genome, code for proline-rich proteins of 31 and 16 kDa, respectively, which show a distant relationship with the putative movement proteins (ORF2) of tymoviruses. GFkV is restricted to Vitis species, which are infected latently, with the exception of V. rupestris, which reacts to GFkV with translucent spots (flecks) on the leaves. GFkV is strictly confined to the phloem of infected hosts and is not transmissible by sap inoculation. The cytopathology of GFkV infections is characterized by a severe modification of mitochondria into structures called “multivesiculate bodies”. Field spread of GFkV has been reported, but the vector is unknown. GFkV is not transmitted through seeds, thus virus dissemination is primarily through distribution of infected propagative material.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Overall genome sequence identity of less than 80%.
- Capsid protein sequences identity less than 90%.
- Serological specificity.
List of species in the genus Maculavirus
| Grapevine fleck virus |
|
|
| Grapevine fleck virus - Italy | [AJ309022 = NC_003347] | (GFkV-IT) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Maculavirus but have not been approved as species
| Bombyx mori macula-like latent virus | [AB186123] | (BmMLV) |
| Grapevine red globe virus | [AJ249360, AF521977]* | (GRGV) |
* Sequences do not comprise the complete genome.
List of unassigned species in the family Tymoviridae
| Bombyx mori latent virus |
|
|
| Poinsettia mosaic virus | [AJ271595 = NC_002164] | (PnMV) |
List of other related viruses which may be members of the family Tymoviridae but have not been approved as species
| Olive latent virus 3 | [FJ444852 = NC_013920] | (OLV3) |
PnMV has characteristics of both the tymoviruses and marafiviruses, and some distinct characteristics. Like tymoviruses, PnMV is mechanically transmissible, infects all main tissues of the host, and amplifies to high titre. Like that of the marafivirus OBDV, the genome contains a single long ORF comprising fused replication and CP domains, a poly(A) tail at the 3′ end and a “marafibox” variant of the “tymobox”. Unlike marafiviruses, PnMV is not insect-transmitted, and possesses only a single CP. PnMV infection induces invaginations of the chloroplast membrane, but these are bound by a single membrane, rather than the double membrane, characteristic of tymoviruses. Sequence relationships (Figure 5) support classification distinct from the existing genera of the family Tymoviridae.
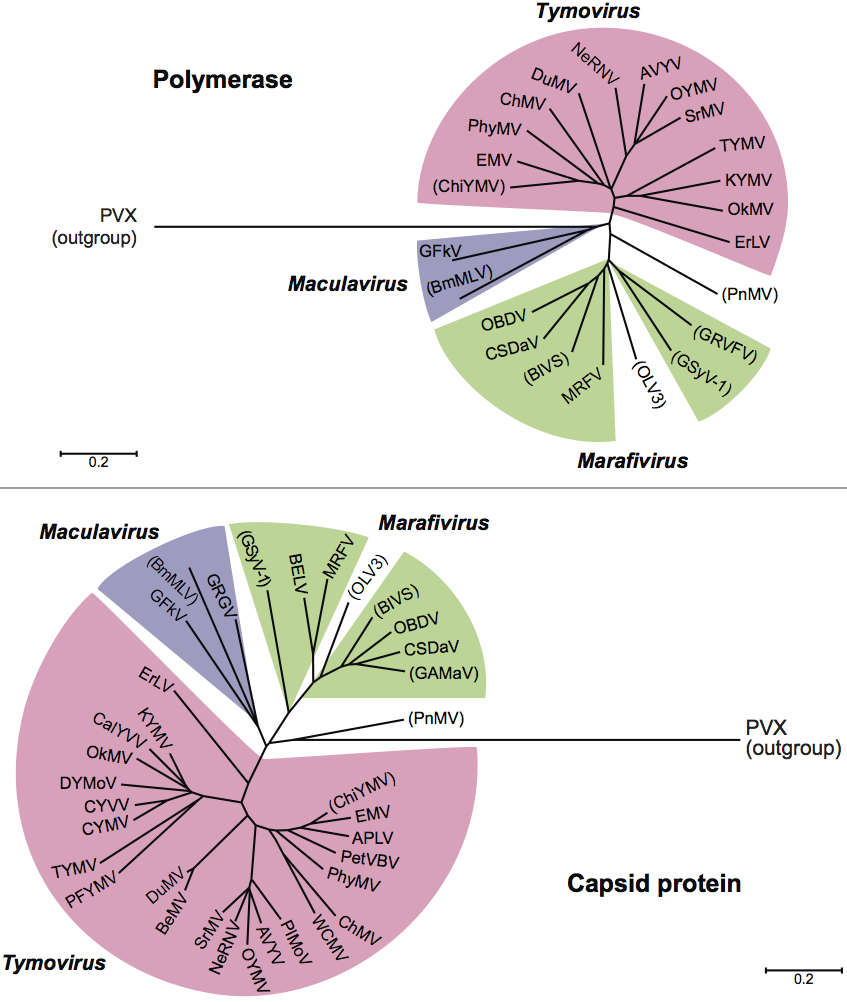
Phylogenetic relationships within the family
Phylogenetic relationships within the family are depicted in Figure 5.
Similarity with other taxa
The replication polyproteins are members of the “alphavirus-like” supergroup of RNA viruses and are most closely related to those of the other families in the order, namely Alphaflexiviridae, Betaflexiviridae and Gammaflexiviridae.
Derivation of names
Macula: from macula, Latin for “fleck”.
Marafi: from maize rayado fino virus.
Tymo: from turnip yellow mosaic virus.
Further reading
Bozarth, C.S., Weiland, J.J. and Dreher, T.W. (1992). Expression of ORF-69 of turnip yellow mosaic virus is necessary for viral spread in plants. Virology, 187, 124-130.
Bradel, B.G., Preil, W.X. and Jeske, H. (2000). Sequence analysis and genome organisation of Poinsettia mosaic virus (PnMV) reveal closer relationship to marafiviruses than to tymoviruses. Virology, 271, 289-297.
Canady, M.A., Larson, S.B., Day, J. and McPherson, A. (1996). Crystal structure of turnip yellow mosaic virus. Nature Struct. Biol., 3, 771-781.
Ding, S.W., Howe, J., Keese, P., Mackenzie, A., Meek, D., Osorio-Keese, M., Skotnicki, M., Pattana, S., Torronen, M. and Gibbs, A. (1990). The tymobox, a sequence shared by most tymoviruses: its use in molecular studies of tymoviruses. Nucl. Acids Res., 18, 1181-1187.
Dreher, T. W. and Goodwin, J.B. (1998). Transfer RNA mimicry among tymoviral genomic RNAs ranges from highly efficient to vestigial. Nucl. Acids Res., 26, 4356-4364.
Edwards, M.C. and Weiland, J.J. (2010). First infectious clone of the propagatively transmitted Oat blue dwarf virus. Arch. Virol., 155, 463-70.
Edwards, M.C. and Weiland, J.J. (2011). Presence of a polyA tail at the 3' end of maize rayado fino virus RNA. Arch. Virol., 156, 331-4.
Edwards, M.C., Zhang, Z. and Weiland, J.J. (1997). Oat blue dwarf marafivirus resembles the tymoviruses in sequence, genome organization, and expression strategy. Virology, 232, 217-229.
Hammond, R.W. and Ramirez, P. (2001). Molecular characterization of the genome of Maize rayado fino virus, the type member of the genus Marafivirus. Virology, 282, 338-347.
Hellendoorn, K., Mat, A.W., Gultyaev, A.P. and Pleij, C.W.A. (1996). Secondary structure model of the coat protein gene of turnip yellow mosaic virus RNA: long C-rich, single-stranded regions. Virology, 224, 43-54.
Izadpanah, K., Zhang, Y.P., Daubert, S. and Rowhani, A. (2002). Sequence of the coat protein gene of Bermuda grass etched-line virus and of the adjacent "marafibox" motif. Virus Genes, 24, 131-134.
Jakubiec, A., Drugeon, G., Camborde, L. and Jupin, I. (2007). Proteolytic processing of turnip yellow mosaic virus replication proteins and functional impact on infectivity. J. Virol. 81, 11402-12.
Katsuma, S. et al., (2005). Novel macula-like virus identified in Bombyx mori cultured cells. J. Virol., 79, 5577-84.
Koenig, R. (1976). A loop structure in the serological classification system of tymoviruses. Virology, 72, 1-5.
Koenig, R., Barends, S., Gultyaev, A.P., Lesemann, D.-E., Vetten, H.J., Loss, S and Pleij, C.W.A. (2005). Nemesia ring necrosis virus: a new tymovirus with a genomic RNA having a histidylatable tobamovirus-like 3´ end. J. Gen. Virol., 86, 1827-33.
Lesemann, D.E. (1977). Virus group-specific and virus-specific cytological alterations induced by members of the tymovirus group. Phytopathol. Z., 90, 315-336.
Maccheroni, W. et al. (2005). Identification and genomic characterization of a new virus (Tymoviridae family) associated with Citrus Sudden Death Disease. J. Virol. 79, 3028-37.
Martelli, G.P., Sabanadzovic, S., Abou Ghanem-Sabanadzovic, N. and Saldarelli, P. (2002). Maculavirus, a new genus of plant viruses. Arch. Virol., 147, 1847-1853.
Sabanadzovic, S., Abou Ghanem-Sabanadzovic, N. and Gorbalenya, A.E. (2009). Permutation of the active site of putative RNA-dependent RNA polymerase in a newly identified species of plant alpha-like virus. Virology, 394, 1-7.
Sabanadzovic, S., Abou Ghanem-Sabanadzovic, N., Saldarelli, P. and Martelli, G.P. (2001). Complete nucleotide sequence and genome oganization of Grapevine fleck virus. J. Gen. Virol., 82, 2009-2015.
Tzanetakis, I.E., Tsai, C.H., Martin, R.R. and Dreher, T.W. (2009). A tymovirus with an atypical 3´-UTR illuminates the possibilities for 3´-UTR evolution. Virology, 392, 238-45.
Weiland, J.J. and Dreher, T.W. (1989). Infectious TYMV RNA from cloned cDNA. Effects in vitro and in vivo of point substitutions in the initiation codons of two extensively overlapping ORFs. Nucl. Acids Res., 17, 4675-4687.
Supplementary reading
A supplementary list of notable Tymoviridae papers is available online on Science Direct®, www.sciencedirect.com.
Contributed by
Dreher, T.W., Edwards, M.C., Gibbs, A.J., Haenni, A.-L., Hammond, R.W., Jupin, I., Koenig, R., Sabanadzovic, S. and Martelli, G.P.
Figures
Figure 1 (Left) Atomic rendering of a virion of turnip yellow mosaic virus (TYMV) (Canady et al., Nature Structural Biology, 3, 771781, 1996). (Center) Diagram of a TYMV virion with CP clusters in hexa- and pentamers. (Right) Negative contrast electron micrograph of virions and empty particles of Belladonna mottle virus (BeMV), a representative of the genus Tymovirus. Particles of members of the genera Marafivirus and Maculavirus have the same morphology. The bar represents 100 nm.
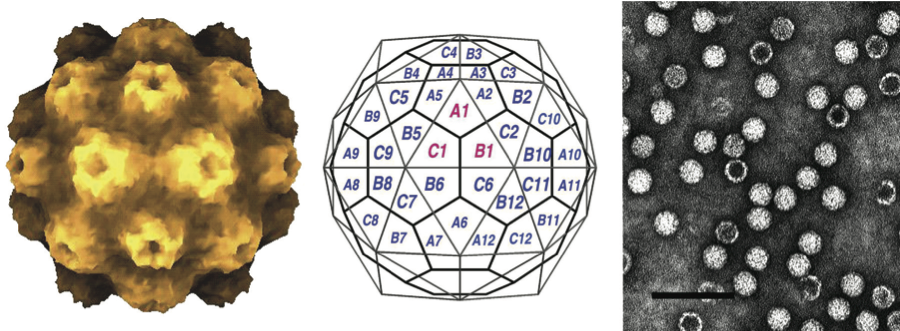
Figure 2 Organization and expression of the genome of turnip yellow mosaic virus (TYMV). Mtr, methyltransferase; Pro, papain-like protease; Hel, helicase; Pol, polymerase (RdRp).
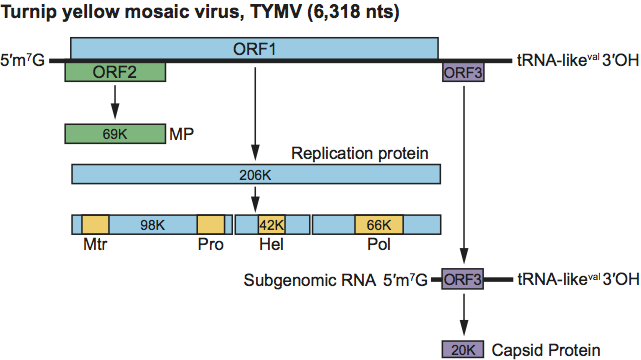
Figure 3 The two known types of genome structure in the genus Marafivirus, exemplified by maize rayado fino virus (MRFV) and oat blue dwarf virus (OBDV), showing the relative position of the ORFs and their expression products. Mtr, methyltransferase; Pro, papain-like protease; Hel, helicase; Pol, polymerase (RdRp); CPs, capsid proteins; p43, proline-rich protein.
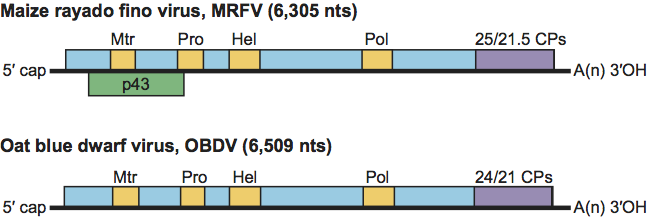
Figure 4 Genome structure of an isolate of Grapevine fleck virus, the type species of the genus Maculavirus showing the relative position of the ORFs and their expression products. Mtr, methyltransferase; Pro, papain-like protease; Hel, helicase; Pol, polymerase (RdRp); CP, capsid protein; p31 and p16, proline-rich proteins.
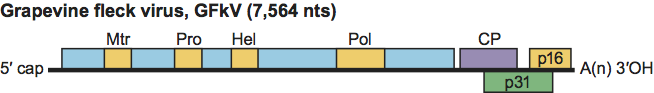
Figure 5 Phylogenetic tree showing the relationship between the species and genera of the family Tymoviridae based on the RdRp (top) and CP (bottom) sequences. The abbreviations and the sequence accession numbers are indicated in the lists of species. The tree was produced using the neighbor-joining method in MEGA4 and is drawn to scale. Names enclosed in parentheses denote viruses that have not yet been approved as species belonging to the Tymoviridae. Note that OLV-3, which has sequences related to the marafiviruses, possesses a maculavirus-like genome organization, and is therefore likely to be proposed as an unassigned species in the family Tymoviridae.