Order: Tymovirales
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Introduction
The order Tymovirales contains viruses that mostly infect plants, have a single molecule of positive sense ssRNA and which are united by the similarities in their replication-associated polyproteins (which account for the majority of the coding capacity of the genome). While there are differences in particle morphology between families, phylogenetic analysis requires the family Tymoviridae (with isometric virions) to be included within a “flexivirus” grouping of viruses that have filamentous virions. There is therefore a convincing case for a common ancestor for all members of the Tymovirales that excludes all other viruses so far characterized.
Virion properties
Morphology
Virions within the families Alphaflexiviridae, Betaflexiviridae and Gammaflexiviridae are flexuous filaments, usually 12–13 nm in diameter and from about 470 to 1000 nm in length, depending on the genus. They have helical symmetry and in some genera there is clearly visible cross-banding. Members of the family Tymoviridae have isometric virions about 30 nm in diameter with a rounded contour and prominent surface structure.
Physicochemical and physical properties
These vary, depending on the particle morphology. Those with filamentous particles usually sediment as single bands with an S20,w of 92–176S, depending on the genus. In the family Tymoviridae, particles sediment as two centrifugal components (S20,w values of 42–55 and 109–125S).
Nucleic acid
In all members of the order, virions contain a single molecule of positive sense ssRNA between 5.9 and 9.0 kb in length. This constitutes 5–6% by weight of the virion in those with filamentous particles and 25–35% in those with isometric virions (Tymoviridae). The genomes are polyadenylated at the 3’ terminus (except in the genus Tymovirus where they usually have a tRNA-like structure at the 3’ end) and capped with m7G at the 5’ end.
Proteins
Almost all members have a single coat protein of 18–44 kDa. In a few instances (genus Lolavirus and some marafiviruses) there are two structural proteins which are different forms from the same gene. In a single case (genus Sclerodarnavirus) no structural protein is known.
Lipids
None reported.
Carbohydrates
Usually none but there are reports that the coat protein is glycosylated in Lolium latent virus (genus Lolavirus) and in some strains of potato virus X (genus Potexvirus).
Genome organization and replication
The largest protein encoded is a replication-associated polyprotein of about 150–250 kDa close to the 5’ end of the genome and which is translated directly from the genomic RNA. The protein contains a set of functional domains whose amino acid sequences and order are conserved in all viruses of the alphavirus-like superfamily of positive-strand RNA viruses. A methyltransferase type 1 domain (Mtr) is located near the N terminus and an RNA-dependent RNA polymerase domain (RdRp) with a characteristic core motif S/TGx3Tx3NS/Tx22GDD occurs near the C terminus. An RNA helicase domain of superfamily 1 (Hel) is localized upstream from the RdRp domain. The Mtr is involved in capping the RNA, the Hel in unwinding RNA and the RdRp in RNA synthesis. Upstream from the Hel in some genera, there is a papain-like cysteine protease domain (P-Pro) that processes the replication polyprotein. Immediately upstream of the P-Pro, some members have a protease of the Ovarian Tumor (OTU) family. Finally, some members have an AlkB (alkylated DNA repair protein) motif between the Mtr and protease or helicase regions. Except in the genus Sclerodarnavirus, where the replicase is the only protein, there are 1 to 5 smaller proteins, translated in most cases from subgenomic RNAs. These include the capsid protein and one or more proteins involved in cell-to-cell movement. The exact organization of the genome is usually a characteristic of the individual genus.
Antigenic properties
These vary depending on the genus.
Biological properties
Most members of the order infect plants but a few species are from plant pathogenic fungi. Host range and transmission are often characteristic of individual genera.
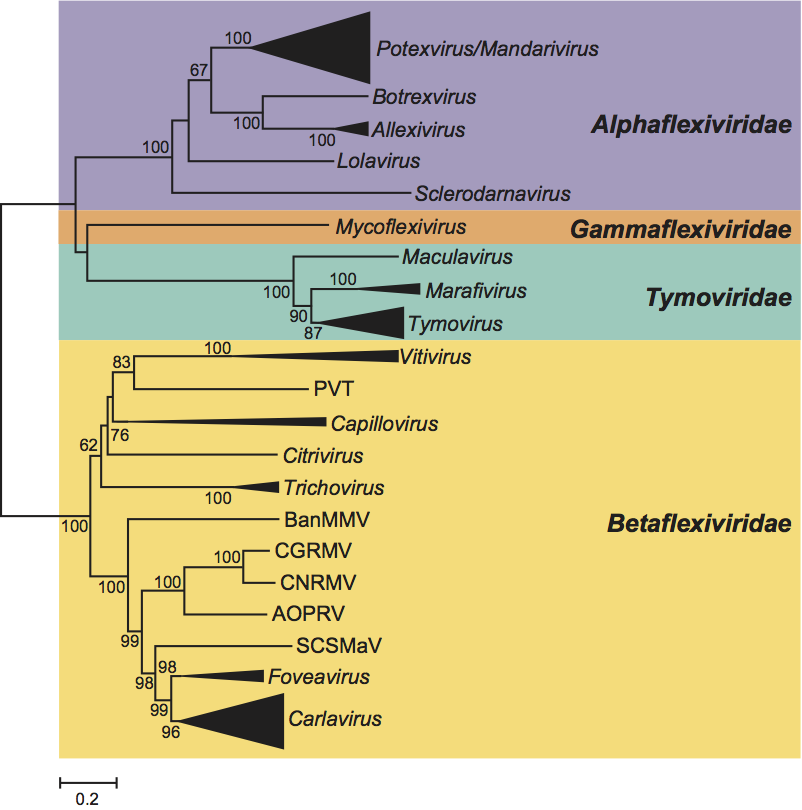
Phylogenetic relationships within the order
In phylogenetic analysis of the replication protein, each genus and family forms a distinct, well-supported branch. The viruses with flexuous virions in the families Alphaflexiviridae and Gammaflexiviridae are more closely related to those with icosohedral particles in the family Tymoviridae than they are to members of the Betaflexiviridae (Figure 1).
Figure 1 Phylogenetic (distance) tree based on the amino acid sequences of the entire replication protein of members of the order Tymovirales. A single representative isolate of each sequenced species in the order was included. Genera and families (which are all monophyletic) have been collapsed into a triangle, the size of which corresponds to the variation found within the clade. Numbers on branches indicate percentage of bootstrap support out of 1000 bootstrap replications (when >60%). The scale indicates JTT (Jones-Taylor-Thornton matrix) amino acid distances. Tree produced in MEGA4.
Similarity with other taxa
In an analysis of the replication protein, the order forms a very distinct group within the alphavirus-like superfamily, with a rather distant relationship to plant virus families such as Closteroviridae, Bromoviridae and Virgaviridae.
Derivation of name
The name is derived from the genus Tymovirus (and family Tymoviridae). This was chosen because the other constituent families have names reflecting their flexuous virions (not a characteristic of all members of the order).
Further reading
Martelli, G., Adams, M.J., Kreuze, J.F., Dolja, V.V., 2007. Family Flexiviridae: a case study in virion and genome plasticity. Ann. Rev. Phytopathol. 45, 73–100.
Contributed by
Adams, M.J., Kreuze, J.F. and Martelli, G.P.
