Family: Tombusviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Capsids exhibit T=3 icosahedral symmetry and are composed of 180 identical protein subunits in three conformationally distinct states (A,B,C) (Figure 1). Capsids are formed with CP having one of two distinct phylogenetic origins. The virions from the genera Aureusvirus, Avenavirus, Carmovirus, Dianthovirus and Tombusvirus have a rounded outline, a granular surface and a diameter of about 32–35 nm. Each subunit folds into three distinct structural domains: R, the N-terminal internal domain interacting with RNA; S, the shell domain constituting the capsid backbone; and P, the protruding C-terminal domain, which gives the virus its granular appearance. P domains are clustered in pairs to form 90 projections. The S domain forms a beta barrel structure made up of eight β-strands. Two Ca2+ binding sites stabilize contacts between adjacent S domains of the A, B and C subunits at the pseudo three-fold axis. The capsids of viruses in the genera Machlomovirus, Necrovirus and Panicovirus are composed of similarly structured CPs that lack the P domain and thus have a smooth appearance. Particles range in diameter between 30–32 nm, and the S domain is related to the CPs of sobemoviruses.
Physicochemical and physical properties
Virions sediment as one component with an S20,w of 118–140S, have a buoyant density ranging from 1.34 to 1.36 g cm−3 in CsCl, and a virion Mr of 8.2–8.9×106. Virions are stable at acidic pH, but expand above pH 7 and in the presence of EDTA. Virions are resistant to elevated temperatures (thermal inactivation usually occurs above 80 °C) and are insensitive to organic solvents and non-ionic detergents.
Nucleic acid
With the exception of those of the genus Dianthovirus, virions contain a single molecule of positive sense, linear ssRNA, that constitutes about 17% of the particle weight, and have a size ranging from 3.7 to 4.8 kb, depending on the genus. Dianthovirus virions contain two genomic RNAs of approximately 3.8 kb and 1.4 kb. The 5’-end of the genome is uncapped. The 3’-ends are not polyadenylated. dsRNAs corresponding in size to viral genomic and sgRNAs are present in infected tissues. DI RNAs occur in some genera. In addition, some members have satellite RNAs or satellite viruses associated with them.
Proteins
Capsids are composed of one of two phylogenetically distinct groups of CP subunit. In those genera that have a CP lacking a protruding domain, the CP subunit ranges in size from 25–30 kDa. In those genera with a CP containing a P domain, the CP subunit ranges in size from 37 to 48 kDa.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Even though variability exists in the number and location of genes within members of the family, a number of organizational features are highly conserved (Figure 2). The unifying feature of the family is that each member species possesses a highly conserved polymerase that is interrupted by an in-frame termination codon that is periodically suppressed to express the core polymerase containing the canonical “GDD” motif. Dianthoviruses utilize a −1 ribosomal frameshifting mechanism to accomplish the same result. The polymerase is further characterized by possessing no obvious helicase or methlytransferase motifs. In this description, as well as those for each genus, the polymerase is labeled as a single ORF with the readthrough (RT) portion labeled ORF1-RT or ORF1-FS.
Genomes of members of the genera Dianthovirus and Avenavirus encode three ORFs, members of the genus Necrovirus and Panicovirus encode five ORFs, while all others encode four ORFs. In the genus Machlomovirus there is an additional putative terminator readthrough event to express an accessory ORF (Figure 2, ORF3-RT). Products of the 5’-proximal ORFs 1 and 1RT or 1FS are expressed by translation directly from the genomic RNA, whereas translation products of the internal and 3’-proximal ORFs are expressed from subgenomic RNAs (sgRNAs). Translation of the genome is cap-independent and is controlled by elements in the 3′-terminal untranslated region of the genome. These elements form base pairs with sequences in the 5′-terminal region of the template RNA, enhancing translation of RNA. For all genera, the CP ORF is either internal or 3′-proximal and requires the synthesis of a sgRNA for expression in vivo. In most cases, it has been demonstrated that the CP is not required for cell-to-cell movement but is required for or facilitates long distance movement.
Non-structural proteins include the phylogenetically conserved pre-readthrough proteins of 23–48 kDa and its 82–112 kDa readthrough products. Tombusviruses and aureusviruses encode a conserved 14–19 kDa protein (see Figure S2) which is associated with the suppression of silencing. Viruses in the family utilize at least four phylogenetically distinct MPs. Genomes of tombusviruses and aureusviruses encode a conserved 22–27 kDa MP (see Figure S1) and the dianthovirus genome encodes a second type of MP of about 34 kDa. The carmoviruses, machlomoviruses, necroviruses and panicoviruses encode a 7–8 kDa MP (MP1) which contains a conserved carboxyl-terminus (see Figure S3). The carmoviruses, necroviruses and panicoviruses encode an additional conserved small 6–9 kDa polypeptide (MP2) (see Figure S4). Panicoviruses and machlomoviruses have additional accessory ORFs whose functions have not been determined.
Replication occurs in the cytoplasm, in membranous vesicles that may be associated with endoplasmic reticulum, or modified organelles such as peroxisomes, mitochondria and, more rarely, chloroplasts. Virions are present in the cytoplasm and occasionally in mitochondria and nuclei.
Antigenic properties
Virions are efficient immunogens. Antisera yield single precipitin lines in immunodiffusion tests. Depending on the genus, serological cross-reactivity among species ranges from nil to near-homologous titers. Many serologically related strains have been identified in several species.
Biological properties
Host range
The natural host range of individual virus species is relatively narrow. Members can either infect monocotyledonous or dicotyledonous plants, but no species can infect both. The experimental host range is wide. Infection is often limited to the root system, but when hosts are invaded systemically, viruses enter all tissues. Many members induce a necrosis symptom in the foliar parts of the plant. Diseases are characterized by mottling, crinkling, necrosis and deformation of foliage. Some infections are symptomless in their natural hosts.
Transmission
All members are readily transmitted by mechanical inoculation and through plant material used for propagation. Some may be transmitted by contact and through seeds. Viruses are often found in natural environments, i.e. surface waters and soils from which they can be acquired without assistance of vectors. Transmission by the chytrid fungus in the genus Olpidium and by beetles has also been reported for members of several genera. Most members can be transmitted through the soil either dependent on, or independent of, a biological vector.
Geographical distribution
Geographical distribution of particular species varies from wide to restricted. The majority of the species occur in temperate regions. Legume-infecting carmoviruses and one possible member of the genus Dianthovirus have been recorded from tropical areas.
Cytopathic effects
Distinctive cytopathological features occur in association with exceedingly high accumulations of virus particles in cells and “multivesicular bodies”, i.e. cytoplasmic membranous inclusions originated from profoundly modified mitochondria and/or peroxisomes. Dense granules which may consist of coat protein aggregates are also found in infected cells.
Genus demarcation criteria in the family
The list of criteria demarcating genera in the family are:
- Structural criteria: spherical virions with either a smooth or granular appearance.
- Genomic criteria: genome organization, number of genome segments, size of genome.
- Polymerase criteria: gene interrupted by a termination codon or a −1 ribosomal frameshifting element that is periodically read through.
Genus Tombusvirus
Type species Tomato bushy stunt virus
Distinguishing features
The genome is approximately 4.7–4.8 kb and contains four ORFs. The CP ORF is located internally on the genomic RNA and is expressed in vivo from an approximate 2.2 kb sgRNA. ORFs 3 and 4 are 3’ proximally located and ORF4 is contained within ORF3 in a different reading frame. Both ORFs are expressed from a second sgRNA of about 0.9 kb. The genome organization and expression strategy are identical to that of aureusviruses. The tombusvirus ORF3 is significantly smaller and ORF4 significantly larger than that in the aureusviruses. The ORF3 MP shares sequence similarity with that of the aureusviruses (see Figure S1) whereas no sequence similarity is observed with other members of the family Tombusviridae. All members elicit formation of multivesicular inclusion bodies. Diseases caused by tombusviruses prevail in temperate climates. All species are soil-borne, but only one, cucumber necrosis virus (CNV), has a recognized fungal vector (Olpidium bornovanus).
Virion properties
Morphology
Capsids are 32–35 nm T=3 icosahedra and have the structure described above for family members containing a P domain. Recent cryo-electron microscopy studies show that the CNV particle consists of a structured inner shell that forms conspicuous connections with the outer shell at the particle three-fold axis (Figure 3). Viral RNA lies along the inner face of the outer shell and may also extend into the inner shell. The structured inner shell may serve as a scaffold for assembly of the particle.
Physicochemical and physical properties
The virus sediments as one component with an S20,w of 132–140S, has a buoyant density of 1.34–1.36 g cm−3 in CsCl, and a virion Mr of 8.9×106. The virion isoelectric point is pH 4.1. Particles exhibit an A260/A280 ratio of 1.64 and a thermal inactivation point of 80–90 °C. Longevity in vitro of 130–150 days has been reported. Virions have a dilution end point in excess of 10−6. Virions are insensitive to ether, chloroform and non-ionic detergents. Virions are stabilized by divalent cations.
Nucleic acid
Nucleic acid represents 17% of the virion, and consists of one molecule of linear positive sense ssRNA. Total genome length is approximately 4.8 kb. The 3′ terminus has neither a poly(A) tract nor a terminal tRNA-like structure. A 3’-proximal segment is involved in facilitating cap-independent translation. In addition to genomic RNA, virions of some species harbor and package DI and/or satellite RNAs. sgRNAs may also be packaged into virions at various levels. sgRNAs are generated by premature termination during genome minus strand synthesis, followed by sgRNA production using the truncated minus strand RNA as template. Three virus-specific dsRNA species are found in infected cells. These correspond to the genomic RNA and the two sgRNAs.
Genome organization and replication
The genomic RNA contains four ORFs (Figure 4). ORF1 encodes a 32–36 kDa protein. This protein has transmembrane binding regions for anchoring to peroxisomal or mitochondrial outer membranes and is involved in recruiting viral RNA to the site of replication. Readthrough (RT) of the ORF1 amber termination codon allows expression of a 92–95 kDa protein (ORF1-RT). Both the 32–36 and 92–95 kDa proteins are produced by translation of genomic RNA. The ORF1 and ORF1-RT-encoded proteins comprise part of the viral RNA-dependent RNA polymerase (RdRp) which is conserved among members of the Tombusviridae (see Figure S6). The polymerase is also distantly related to the flavivirus and pestivirus polymerases (see Figure S6). The internal ORF2 encodes the CP, which is expressed from the approximate 2.2 kb sgRNA1. The CP is less conserved than the polymerase. Two nested ORFs (ORF3 and ORF4) located at the 3’ terminus of the genome, encode proteins of about 22 kDa (p22) and about 19 kDa (p19), respectively. p22 is required for cell-to-cell movement, interacting with a host homeodomain leucine-zipper protein. p19 is a suppressor of RNA silencing. It also has a role in the systemic spread of the virus, and is involved in host specific development of necrosis. p22 and p19 are related to the ORF 4 and 5 proteins of aureusviruses (see Figures S1 and S2) and are not related to proteins encoded by other members of the Tombusviridae.
Genome replication is carried out by the virus encoded polymerase in conjunction with at least eight host factors. The replication process begins with the synthesis of a minus-strand RNA from a plus-strand template. The minus-strand is then used as template for the synthesis of the progeny genomes. Numerous long-range RNA/RNA interactions regulate viral RNA replication, transcription and translation. Cap-independent translation utilizes a Y-shaped structure in the 3′ region of the genome that base-pairs with sequences in the 5′ end of the genome. DI RNAs are generated during replication of some species following multiple and progressive deletions of genomic RNA. Cymbidium ringspot virus (CymRSV), carnation Italian ringspot virus (CIRV) and TBSV RNA can replicate in cells of the yeast Saccharomyces cerevisiae. Over 150 host proteins have been found to affect the efficiency of viral RNA replication in yeast. DI RNAs are also replicated in yeast.
Antigenic properties
Most species are serologically interrelated, though to a variable extent. Species with serologically related virions are: artichoke mottled crinkle virus, pelargonium leaf curl virus and petunia asteroid mosaic virus (PAMV), which are closely related; Moroccan pepper virus (MPV) which is distantly related; eggplant mottled crinkle virus (EMCV), CIRV, Lato river virus and Neckar river virus (NRV) which are very distantly related. Species with serologically unrelated virions are CymRSV and CNV.
Biological properties
Host range
Most species have a narrow natural host range. However, most also have a wide experimental host range. Even though the host range of an individual species is restricted in nature, tombusviruses are present in a wide range of both monocotyledonous and dicotyledonous plants. Viruses tend to remain localized, forming a necrosis in artificially infected hosts. However, Nicotiana clevelandii and N. benthamiana are often systemically infected.
Transmission
Members are readily transmitted mechanically in the field and experimentally. Most, if not all, species are soil-borne without the aid of a biological vector. They appear to be directly transmitted through the soil. CNV is transmitted by the chytrid fungus Olpidium bornovanus. Some species may be transmitted through the seed at a very low level.
Geographical distribution
TBSV strains are present throughout North and South America, Europe, and the Mediterranean. Other tombusviruses are present wherever the primary hosts exist.
Cytopathic effects
Virions are found in all parts of the host plant cells including cytoplasm, nuclei, nucleoli, mitochondria and cell vacuoles. Virus crystals are present in the cytoplasm of infected cells. Multivesicular bodies (MVBs), i.e. cytopathic structures made up of a main body surrounded by spherical to ovoid vesicles 80–150 nm in diameter, are consistently present in infected cells. MVBs originate from the proliferation of the limiting membrane of peroxisomes (e.g. CymRSV, TBSV and CNV) or mitochondria (CIRV), their formation being determined by an ORF1 encoded sequence as short as about 200 aa. MVBs do not contain virions. However, virions are present within seemingly intact mitochondria in cells infected by EMCV, CymRSV, grapevine Algerian latent virus, MPV, NRV, TBSV and occasionally PAMV.
Species demarcation criteria in the genus
The list of species demarcation criteria in the genus is:
- Extent of serological relationship as determined by immunodiffusion (usually not below 3), and/or ELISA
- Less than 85% sequence identity in the CP
- Differential cytopathological features; organelles from which MVBs arise
- Natural host range
- Artificial host range reactions
List of species in the genus Tombusvirus
|
Artichoke mottled crinkle virus |
|
|
|
Artichoke mottled crinkle virus - Bari |
[X62493=NC_001339] |
(AMCV-Bari) |
|
Carnation Italian ringspot virus |
|
|
|
Carnation Italian ringspot virus - Italy |
[X85215=NC_003500] |
(CIRV-IT) |
|
Cucumber Bulgarian latent virus |
|
|
|
Cucumber Bulgarian latent virus - Bulg |
[AY163842=NC_004725] |
(CBLV-Bulg) |
|
Cucumber necrosis virus |
|
|
|
Cucumber necrosis virus - Canada |
[M25270=NC_001469] |
(CNV-Can) |
|
Cymbidium ringspot virus |
|
|
|
Cymbidium ringspot virus - UK |
[X15511=NC_003532] |
(CymRSV-UK) |
|
Eggplant mottled crinkle virus |
|
|
|
Eggplant mottled crinkle virus - Israel |
[FJ977166*] |
(EMCV-Is) |
|
Lisianthus necrosis virus - Taiwan |
[DQ011234=NC_007983] |
(LNV-TW) |
|
Pear latent virus - Italy |
[AY100482=NC_004723] |
(PLV-IT) |
|
Grapevine Algerian latent virus |
|
|
|
Grapevine Algerian latent virus - Japan nipplefruit |
[AY830918=NC_011535] |
(GALV-JA) |
|
Havel river virus |
|
|
|
Havel river virus - S |
[AY370535*] |
(HRV-S) |
|
Lato river virus |
|
|
|
Lato river virus - Italy |
|
(LRV-IT) |
|
Limonium flower distortion virus |
|
|
|
Limonium flower distortion virus - Lim2 |
[AY500882*] |
(LFDV-Lim2) |
|
Moroccan pepper virus |
|
|
|
Moroccan pepper virus - Italy |
[AF540886*] |
(MPV-IT) |
|
Neckar river virus |
|
|
|
Neckar river virus - Germany |
[AY500887*] |
(NRV-GR) |
|
Pelargonium leaf curl virus |
|
|
|
Pelargonium leaf curl virus - Koenig |
[AF290026*] |
(PLCV-Koenig) |
|
Pelargonium necrotic spot virus |
|
|
|
Pelargonium necrotic spot virus - UPEV |
[AJ607402=NC_005285] |
(PNSV-UPEV) |
|
Petunia asteroid mosaic virus |
|
|
|
Petunia asteroid mosaic virus - Lim1 |
[AY500881*] |
(PAMV-Lim1) |
|
Sikte waterborne virus |
|
|
|
Sikte waterborne virus - Lim6 |
[AY500886*] |
(SWBV-Lim6) |
|
Tomato bushy stunt virus |
|
|
|
Tomato bushy stunt virus - cherry |
[M21958=NC_001554] |
(TBSV-cherry) |
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Tombusvirus but have not been approved as species
|
Lettuce necrotic stunt virus |
[AJ288944] |
(LNSV) |
Genus Dianthovirus
Type species Carnation ringspot virus
Distinguishing features
Virions sediment in sucrose gradients as a single band of S20,w 126–135S. The genome is composed of two RNAs of approximately 3.8 and 1.4 kb (Figure 6). ORF1 encodes the polymerase and is interrupted by a ribosomal frameshifting event yielding a pre-frameshift approximate 27 kDa protein and an approximate 88 kDa frameshift polypeptide. The CP, possessing a protruding domain, is encoded by the 3′-proximal ORF on RNA1 and is expressed from an approximate 1.5 kb sgRNA in vivo. The ORF for the 34–35 kDa MP is in the monocistronic RNA2. The movement protein aa sequence is unique to the dianthoviruses. Members are transmitted through the soil without the aid of a biological vector.
Virion properties
Morphology
Capsids are approximately 37 nm in diameter and have T=3 icosahedral symmetry (Figure 5) as described above for family members containing a P domain. Cryo-electron microscopy has revealed that red clover necrotic mosaic virus (RCNMV) capsids contain a defined inner shell (Figure 5), consisting of a cage containing complexes of virion RNA and the N-terminal R-domain region of the CP. Virions undergo significant structural changes upon removal of the Ca2+ and Mg2+ ions that are found in the shell and interior of the capsid. Loss of these ions in the divalent cation depleted cytoplasm of cells may contribute to particle disassembly.
Physicochemical and physical properties
Virions sediment as one component in sucrose with S20,w of 126–135S. The buoyant density in CsCl is 1.363–1.366 g cm−3, and the virion Mr is 8.6×106. Particles exhibit an A260/A280 ratio of 1.67 and a thermal inactivation point between 80–90 °C. A longevity in vitro of around 10 weeks has been reported for most members. Virions are insensitive to ether, chloroform and non-ionic detergents. Virions are stable at pH 6 and lower; alkaline conditions (pH 7–8) induce particle swelling. Virions are stabilized by divalent cations.
Nucleic acid
Virions contain two molecules of infectious linear positive sense ssRNA of 3.8 and 1.4 kb. The two genomic components are not capped. They do not contain a 3’-terminal poly(A) tract. Virions package either both genomic RNAs or multiple copies of RNA-2. Three virus-specific dsRNA species are found in infected cells. The largest and smallest dsRNAs correspond to the genomic RNA1 and RNA2, respectively. The intermediate sized dsRNA corresponds to the 1.5 kb sgRNA.
Genome organization and replication
RNA1 contains two ORFs. ORF1 is capable of encoding a 27 kDa protein. A −1 ribosomal frameshift event at the canonical shifty heptanucleotide allows translation to continue into ORF1-FS to produce the 88 kDa protein. The ORF1 and ORF1-FS encoded proteins form the viral polymerase. ORF2 encodes the 37–38 kDa CP. This ORF is expressed in vivo from the approximately 1.5 kb sgRNA. ORF3 on RNA2 encodes the 34–35 kDa MP. The RCNMV MP also serves as a silencing suppressor. For the sgRNA of RNA1 to be expressed, the loop of a stem-loop in RNA2 must base pair within the RNA1 sgRNA promoter. This same stem-loop interaction allows RNA-1 to become encapsidated since RNA-2 contains the origin of assembly. Translation of RNA1 occurs in a cap-independent fashion requiring an element in the 3′ UTR that is similar to the barley yellow dwarf virus (BYDV) translational enhancer.
Antigenic properties
Virus particles are moderately to highly immunogenic. Various serologically distinct strains have been identified. Antisera yield a single precipitin line in agar gel-diffusion assays. Monoclonal antibodies have been identified that cross-react between species.
Biological properties
Host range
In nature, dianthoviruses have moderately broad natural host ranges restricted to dicotyledonous plants. In the laboratory, the experimental host range is much broader, and includes a wide range of herbaceous species in the families Solanaceae, Leguminosae, Cucurbitaceae and Compositae. Most plants are infected locally (non-systemically).
Transmission
The viruses are readily transmitted by mechanical inoculation; they are not known to be seed-transmitted. The viruses are not transmitted by insects, nematodes, or soil-inhabiting fungi, although there is a report that RCNMV is transmitted by Olpidium. However, viruses are readily transmitted through the soil without the aid of a biological vector.
Geographic distribution
Dianthoviruses are widespread throughout the temperate regions of the world.
Cytopathic effects
None reported.
Species demarcation criteria in the genus
The list of species demarcation criteria in the genus is:
- Extent of serological relationship as determined by immunodiffusion and/or ELISA
- Extent of sequence identity between relevant gene products
- Less than 93% aa sequence identity in the polymerase
- Less than 75% aa sequence identity in the CP
- Ability to form pseudorecombinants with the two RNA components
- Transmission through the soil
- Natural host range
- Artificial host range reactions
List of species in the genus Dianthovirus
|
|
RNA1 |
RNA2 |
|
|
Carnation ringspot virus |
|
|
|
|
Carnation ringspot virus - Lommel |
[L18870=NC_003530] |
[M88589=NC_003531] |
(CRSV-Lommel) |
|
Red clover necrotic mosaic virus |
|
|
|
|
Red clover necrotic mosaic virus - Australia |
[J04357=NC_003756] |
[X08021=NC_003775] |
(RCNMV-AUS) |
|
Sweet clover necrotic mosaic virus |
|
|
|
|
Sweet clover necrotic mosaic virus - 59 |
[L07884=NC_003806] |
[S46028=NC_003807] |
(SCNMV-59) |
List of other related viruses which may be members of the genus Dianthovirus but have not been approved as species
|
|
RNA1 |
RNA2 |
|
|
Rice virus X |
[AB033715] |
|
(RVX) |
Genus Aureusvirus
Type species Pothos latent virus
Distinguishing features
The virion is a 30 nm icosahedron that packages the genomic RNA of about 4.2–4.4 kb. The genome contains four ORFs. The CP ORF is located internally in the genomic RNA and is expressed in vivo from a sgRNA of about 2 kb. ORFs 3 and 4 are 3’-proximally located and ORF4 is nested within ORF3 in a different reading frame. Both ORFs are expressed from an approximate 0.8 kb sgRNA. The genome organization and expression strategy are identical to that of the tombusviruses. The polymerase and pre-readthrough regions of aureusviruses are most similar to those of tombusviruses (see Figure S6). The aureusvirus ORF3 is significantly larger and ORF4 is significantly smaller than those in the tombusviruses but their encoded proteins are similar to the corresponding ORFs of tombusviruses (Figures S1 and S2). Transmission occurs through the soil without (pothos latent virus; PoLV) or with (cucumber leaf spot virus; CLSV) the aid of a vector (Olpidium bornovanus).
Virion properties
Morphology
Virions are isometric with a rounded outline, a knobby surface and a diameter of about 30 nm (Figure 7). Based on comparative aa sequence alignments, the CP subunits likely contain the R, S and P domain structures described for TBSV and RCNMV. Similarly, particles are likely T=3 icosahedra composed of 180 copies of the CP.
Physicochemical and physical properties
Preparations of purified virus sediment as a single component in sucrose density gradients, and to equilibrium in solutions of CsCl and Cs2SO4. Buoyant density in CsCl and Cs2SO4 is 1.34–1.36 and 1.37 g cm−3, respectively. The thermal inactivation point is above 80 °C. Virus particles resist organic solvents but are readily disrupted by SDS.
Nucleic acid
Virions contain a single molecule of linear, uncapped, non-polyadenylated, positive sense ssRNA of about 4.2–4.4 kb constituting 17% of the particle weight. Virions can contain the two sgRNAs. Three dsRNA species corresponding to the full-size genomic RNA and the two sgRNAs can be recovered from infected plants. Maize white line mosaic virus (MWLMV) supports the replication of a satellite virus but satellite RNAs or DI RNAs have not been reported for the other aureusvirus members.
Genome organization and replication
The genome of PoLV contains four ORFs (Figure 8). ORF1 encodes a 25 kDa protein. The readthrough of its amber stop codon results in translation into ORF1-RT yielding an 84 kDa polymerase. ORF2 encodes the 40 kDa CP. ORF3 and 4 are nested in different reading frames and encode 27 and 14 kDa proteins, respectively. The 27 kDa protein is the MP and the 14 kDa protein is responsible for symptom severity and is a suppressor of RNA silencing. Both the MP and the silencing suppressor are related to those of tombusviruses (see Figures S1 and S2). The CP is important in regulating the synthesis of the 14 kDa protein, the excess production of which is lethal to infected plants. Two sgRNAs of 2.0 kb and 0.8 kb give rise to the 40 kDa CP and the 27 kDa and 14 kDa proteins, respectively. Translation of sgRNA2 involves a 3′-proximal enhancer element as found for TBSV but translation of sgRNA1 is controlled by a stem loop structure at its 5′ terminus. Replication may occur in the cytoplasm, possibly in association with nucleus-derived vesicles and vesiculated bodies. Virus particles accumulate in the cytoplasm.
Antigenic properties
Virions are efficient immunogens. Polyclonal antisera yield a single precipitin line in immunodiffusion tests and uniformly decorate virus particles. Distant relationships were found with members of the genera Tombusvirus and Carmovirus.
Biological properties
Host range
Pothos (Scindapsus aureus) and pigeonpea (Cajanus cajan) are the only known natural hosts of PoLV, and cucumber (Cucumis sativus) is the natural host of CLSV. The experimental host range is moderately wide. Localized infections are induced in most hosts, except for Nicotiana benthamiana and N. clevelandii, which are systemically invaded. MWLMV is a soil-borne virus that causes disease on sweet corn and is found mainly in the Great Lakes region of the US and Canada.
Transmission
PoLV and CLSV are readily transmitted by mechanical inoculation. Natural transmission occurs through the soil or the circulating solution in hydroponics. CLSV is transmitted by the soil-inhabiting fungus Olpidium bornovanus and, to a low rate (ca. 1%), through seeds. MWLMV may be transmitted by the protist, Polymyxa graminis.
Geographical distribution
Reported from several European countries, Canada, the US, Jordan and India.
Cytopathic effects
PoLV is very invasive in systemically infected plants and is found in parenchyma and conducting tissues. Virus particles often form intracellular crystalline aggregates. Distinctive cytopathological features are the extensive vesiculation of the nuclear envelope and the single-membrane vesiculated bodies in the cytoplasm. The cytopathology of CLSV infections is characterized by occasional peripheral vesiculation of mitochondria and the nuclear envelope and by the presence of membranous cytoplasmic vesicles with fibrillar content. MWLMV infection is associated with lobate nuclei and particles are found in bleb-like evaginations of the tonoplast. Mitochondria contain electron-dense patches.
Species demarcation criteria in the genus
The list of species demarcation criteria in the genus is:
- Serological specificity (known species are serologically unrelated)
- Extent of sequence identity between relevant gene products
- Less than 40% aa sequence identity of the CP
- Less than 80% aa sequence identity of the polymerase
- Differential cytopathological features
- Transmission by a vector
- Natural host range
- Artificial host range reactions
List of species in the genus Aureusvirus
|
Cucumber leaf spot virus |
|
|
|
Cucumber leaf spot virus - Israel |
[DQ227315=NC_007816] |
(CLSV-IS) |
|
Johnsongrass chlorotic stripe mosaic virus |
|
|
|
Johnsongrass chlorotic stripe mosaic virus - Iran |
[AJ557804=NC_005287] |
(JCSMV-IR) |
|
Maize white line mosaic virus |
|
|
|
Maize white line mosaic virus - US |
[EF589670=NC_009533] |
(MWLMV-US) |
|
Pothos latent virus |
|
|
|
Pothos latent virus - India pigeonpea |
[AJ243370=NC_000939] |
(PoLV-IN) |
List of other related viruses which may be members of the genus Aureusvirus but have not been approved as species
|
Sesame necrotic mosaic virus |
[DQ367845*] |
(SNMV) |
* Sequence does not comprise the complete genome.
Genus Avenavirus
Type species Oat chlorotic stunt virus
Distinguishing features
This monotypic genus is distinguished from other genera in the family Tombusviridae because the CP is significantly larger (48K) than other members with a protruding domain and it shares limited aa sequence identity in the coat protein (11%–28%) The genome organization is unique and is intermediate between the tombusviruses and carmoviruses. The 8K protein that overlaps the CP ORF does not share sequence similarity to the small proteins encoded by other Tombusviridae members.
Virion properties
Morphology
Particles are isometric and approximately 35 nm in diameter. Based on sequence similarity with the CPs of members of the family Tombusviridae that contain a protruding domain, it is assumed that the particle has T=3 icosahedral symmetry and is composed of 180 protein subunits with R, S and P structural domains.
Physicochemical and physical properties
No information.
Nucleic acid
The genome is composed of a single positive sense ssRNA molecule of 4,114 nt. It is not known if the 5’ terminus is capped. The 3’ end does not possess a poly(A) tail. A single sgRNA of 1,772 nt is expressed in infected tissues and is encapsidated at low concentrations within the virions.
Genome organization and replication
The genomic RNA contains three ORFs (Figure 9). ORF1 encodes a 23 kDa protein which can be read through to produce the 84 kDa polymerase. These proteins share sequence similarity with the pre-readthrough and readthrough proteins (polymerases) of other members of the Tombusviridae (Figure S6). ORF2 encodes the 48 kDa CP which also shares sequence similarity with other members of the Tombusviridae (Figure S5). ORF3 is within ORF2, in a different reading frame. An ORF encoding a protein with sequence similarity to the MPs of other Tombusviridae members is lacking. ORF3 encodes a unique 8 kDa polypeptide with unknown function but which may serve as a MP. A single 3′ co-terminal sgRNA is formed that presumably serves as template for the CP.
Antigenic properties
The virus is a moderate immunogen. Antibodies do not cross-react with other icosahedral viruses of oats or with representative members of the genera Carmovirus and Machlomovirus.
Biological properties
Host range
The virus has only been identified and studied in oats (Avena sativa).
Transmission
The virus is readily transmitted mechanically from oat plant to oat plant. Infection patterns in winter oat fields are consistent with the virus being soil-borne, and possibly transmitted by zoosporic fungi.
Geographical distribution
This virus has only been reported in the United Kingdom.
Cytopathic effects
None reported.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Avenavirus
|
Oat chlorotic stunt virus |
|
|
|
Oat chlorotic stunt virus - UK |
[X83964=NC_003633] |
(OCSV-UK) |
Species names are in italic script; isolate names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Avenavirus but have not been approved as species
None reported.
Genus Carmovirus
Type species Carnation mottle virus
Distinguishing features
The genomic RNA is about 4.0 kb in size and contains four ORFs. Translation of the genome yields a polypeptide of about 28 kDa encoded by ORF1 and a polypeptide of about 88 kDa (ORF1RT) originating from readthrough of the amber terminator of ORF1. ORFs 2 and 3 code for two small polypeptides of about 7 kDa and 9 kDa that are involved in cell-to-cell movement. The CP contains a P domain and is encoded by ORF4, which is 3’ co-terminal with genomic RNA. The genome organization is similar to that of necroviruses. The carmovirus CP contains a P-domain which is lacking in necroviruses. The ORFs 2, 3 and 4 polypeptides are translated from two sgRNAs with sizes of about 1.7 and 1.5 kb. Members of different species are not serologically related. Multivesicular bodies are formed only by some viruses. Most species are found in temperate regions. Those infecting legumes are reported from tropical areas. Several species are soil-borne. Melon necrotic spot virus (MNSV), cucumber soil-borne virus (CSBV) and squash necrosis virus (SqNV) are transmitted by Olpidium bornovanus.
Virion properties
Morphology
Virions are 32–35 nm in diameter and have T=3 icosahedral symmetry (Figure 10). The isometric nucleocapsids have an obvious regular surface structure giving them a granular appearance in the electron microscope. As with TBSV, TNV and RCNMV, virions are composed of 180 identical protein subunits that fold into the distinctive R, S and P structural domains. The CP subunit ranges in size from about 35–42 kDa. The crystal structures of the carnation mottle virus (CarMV) and cowpea mottle virus (CPMoV) particle have been determined.
Physicochemical and physical properties
Virions sediment as one component with an S20,w of 118–130S. The buoyant density of virions is 1.33–1.36 g cm−3 in CsCl, and the Mr is 8.2×106. CarMV has an isoelectric point of pH 5.2. Particles exhibit an A260/A280 ratio of 1.48–1.66 and a thermal inactivation point of 95 °C. Longevity in vitroof 395 days has been reported. Virions have dilution end points often in excess of 10−6. Virions are insensitive to ether, chloroform and non-ionic detergents, and are stabilized by divalent cations.
Nucleic acid
Nucleic acid comprises 14% of the virion. Virions contain one molecule of linear positive-sense ssRNA. Total genome length varies between about 3.9 and 4.5 kb. The genome is not capped. The 3′-terminus lacks a poly(A) tract. Generally only the genomic RNA is encapsidated. However, some species also harbor and package DI and/or satellite RNAs and sgRNAs may also be packaged into virions at a very low level. Three virus-specific dsRNA species are found in infected cells. These correspond to the genomic and 2 sgRNAs.
Genome organization and replication
The genomic RNA contains four ORFs (Figure 11). In CarMV, ORF1 encodes a 28 kDa protein which upon readthrough of the amber terminator codon yields an 88 kDa polymerase (see Figure S6). ORFs 2 and 3 encode 7 and 9 kDa proteins. The 7 kDa protein is an RNA binding protein and the 9 kDa protein is an integral membrane protein. These and the similar proteins encoded by other carmoviruses function together in cell-to-cell movement of viral RNA. The p7 protein of carmoviruses (MP1) is highly basic and shares sequence similarity with the small proteins encoded by necroviruses, the panicovirus panicum mosaic virus (PMV) and the machlomovirus maize chlorotic mottle virus (MCMV) (see Figure S3). The p9 protein (MP2) has hydrophobic regions and shares sequence similarity with a second small protein encoded by necroviruses and PMV (see Figure S4). Both ORFs 2 and 3 are thought to be expressed in vivo from the larger 1.7 kb sgRNA1 synthesized in infected cells. The ORF3 initiation codon is in a sub-optimal translational context and the ORF2 initiation codon is in an optimal translational context. Ribosome scanning allows translation of ORF3 from the 1.7 kb sgRNA1. ORF4 encodes the 38 kDa CP which is expressed in vivo from the approximate 1.5 kb sgRNA2. For turnip crinkle virus (TCV), hibiscus chlorotic ringspot virus and pelargonium flower break virus the CP has been shown to also be a silencing suppressor. The 5′ and 3′ terminal regions of TCV RNA contain well-characterized sequences regulating viral RNA synthesis. Sequences in the 3′ terminal UTR fold into a Y-shaped structure that control cap-independent translation likely via base pairing interaction with a loop within ORF1. The proximal positioning of the 3′ translation and replication elements likely controls relative levels of transcription and translation of the same viral RNA template.
Antigenic properties
Virions are efficient immunogens. Polyclonal antisera yield a single precipitin line in immunodiffusion tests. Virus species are not serologically related.
Biological properties
Host range
Most species have a narrow natural host range. However, most also have a wide experimental host range. Even though the host range of an individual species is restricted in nature, different carmoviruses infect a wide range of both monocotyledonous and dicotyledonous plants. Viruses tend to remain localized, forming necrosis in artificially infected hosts.
Transmission
The virus is readily transmitted mechanically, under both experimental and natural conditions. CarMV has spread worldwide by the dispersal of infected carnation cuttings. Some species may be transmitted through seed at a low level. Several viruses are soil-borne, and MNSV, CSBV and SqNV are transmitted by Olpidium bornovanus. CPMoV, bean mild mosaic virus, and TCV are transmitted by beetles (Coleoptera).
Geographical distribution
Probably distributed worldwide. Most species are found in temperate regions of the world. Those infecting legumes are reported from tropical areas.
Cytopathic effects
In systemically infected plants virus particles are found in parenchyma and conducting tissues, sometimes forming intracellular crystalline aggregates. Membranous vesicles are produced from the endoplasmic reticulum. Multivesicular bodies are formed only in cells infected by some viruses.
Species demarcation criteria in the genus
The list of species demarcation criteria in the genus is:
- Extent of serological relationship as determined by immunodiffusion and/or ELISA
- Extent of sequence identity between relevant gene products
- Less than 52% aa sequence identity of the CP
- Less than 57% aa sequence identity of the polymerase
- Cytopathological features. Presence or absence of multivesicular bodies
- Transmission by a fungal vector
- Natural host range
- Artificial host range reactions
List of species in the genus Carmovirus
|
Ahlum waterborne virus |
|
|
|
Ahlum waterborne virus - Germany |
|
(AWBV - GR) |
|
Angelonia flower break virus |
|
|
|
Angelonia flower break virus - Florida |
[DQ219415=NC_007733] |
(AFBV-FL) |
|
Bean mild mosaic virus |
|
|
|
Bean mild mosaic virus - Colombia |
|
(BMMV-Col) |
|
Cardamine chlorotic fleck virus |
|
|
|
Cardamine chlorotic fleck virus - CL |
[L16015=NC_001600] |
(CCFV-CL) |
|
Carnation mottle virus |
|
|
|
Carnation mottle virus - Shanghai |
[AF192772=NC_001265] |
(CarMV-SHA) |
|
Cowpea mottle virus |
|
|
|
Cowpea mottle virus - Nigeria |
[U20976=NC_003535] |
(CPMoV-NG) |
|
Cucumber soil-borne virus |
|
|
|
Cucumber soil-borne virus - Lebanon |
|
(CuSBV-LEB) |
|
Galinsoga mosaic virus |
|
|
|
Galinsoga mosaic virus - Queensland |
[Y13463=NC_001818] |
(GaMV-QLD) |
|
Hibiscus chlorotic ringspot virus |
|
|
|
Hibiscus chlorotic ringspot virus - Singapore |
[X86448=NC_ 003608] |
(HCRSV-SING) |
|
Japanese iris necrotic ring virus |
|
|
|
Japanese iris necrotic ring virus - Japan |
[D86123=NC_002187] |
(JINRV-JA) |
|
Melon necrotic spot virus |
|
|
|
Melon necrotic spot virus - Netherlands |
[M29671=NC_001504] |
(MNSV-Neth) |
|
Pea stem necrosis virus |
|
|
|
Pea stem necrosis virus - Japan |
[AB086951=NC_004995] |
(PSNV-JA) |
|
Pelargonium flower break virus |
|
|
|
Pelargonium flower break virus - MZ10 |
[AJ514833=NC_005286] |
(PFBV-MZ10) |
|
Saguaro cactus virus |
|
|
|
Saguaro cactus virus - Arizona |
[U72332=NC_001780] |
(SgCV-Arizona) |
|
Turnip crinkle virus |
|
|
|
Turnip crinkle virus - UK |
[M22445=NC_003821] |
(TCV-UK) |
|
Weddel waterborne virus |
|
|
|
Weddel waterborne virus - Germany |
|
(WWBV - GR) |
Species names are in italic script; isolate names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Carmovirus but have not been approved as species
|
Blackgram mottle virus |
|
(BMoV) |
|
Calibrachoa mottle virus |
[GQ244431] |
(CMoV) |
|
Elderberry latent virus |
[AY038066*] |
(ElLDV) |
|
Glycine mottle virus |
|
(GMoV) |
|
Narcissus tip necrosis virus |
|
(NTNV) |
|
Nootka lupine vein clearing virus |
[EF207438=NC_009017] |
(NLVCV) |
|
Pelargonium chlorotic ring pattern virus |
[AY038069=NC_005985] |
(PCRPV) |
|
Plantain virus 6 |
|
(PlV-6) |
|
Soybean yellow mottle mosaic virus |
[FJ457015=NC_016643] |
(SYMMV) |
|
Squash necrosis virus |
|
(SqNV) |
|
Tephrosia symptomless virus |
|
(TeSV) |
* Sequence does not comprise the complete genome.
Genus Necrovirus
Type species Tobacco necrosis virus A
Distinguishing features
Virions sediment as a single component with an S20w of 118S. The genomic RNA is about 3.7 kb in size and contains four ORFs. A fifth potential smaller ORF is predicted within the 3′ leader of the type member tobacco necrosis virus-A (TNV-A). The genome organization and expression strategy are similar to members of the genus Carmovirus. Necroviruses have a small CP that forms smooth virions and is phylogenetically related to the sobemovirus CP. This feature distinguishes the necroviruses from carmoviruses, which have a larger CP with a protruding domain. ORFs 2 and 3 encode MPs.
Virion properties
Morphology
Virions are approximately 28 nm in diameter and exhibit T=3 icosahedral symmetry. The particle has a smooth appearance (Figure 12). The crystal structure of TNV-A has been determined at 2.25 angstrom resolution. The tertiary and quaternary structures of TNV-A are most similar to those of sobemoviruses. As with other members of the Tombusviridae, the CP subunit has an internal R domain, a shell that has a beta-barrel structure and subunit contacts at the three-fold axis that are stabilized by Ca2+. Necrovirus particles do not have a protruding domain.
Physicochemical and physical properties
The virus sediments as one component with S20,w of 118S, and has a buoyant density of 1.40 g cm−3 in CsCl. The particle has a Mr of 7.6×106. The thermal inactivation point is between 85 and 95 °C. The virion isoelectric point is pH 4.5. Virions are insensitive to ether, chloroform and non-ionic detergents.
Nucleic acid
Virions contain one molecule of infectious linear positive sense ssRNA. The type species RNA is 3,684 nt. The 5’ end of the RNA is not capped, possessing a ppA… terminus. The RNA does not contain a 3’-terminal poly(A) tract. The virion packages exclusively the genomic RNA. Three virus-specific dsRNA species are found in infected cells. The size of the largest virus-specific dsRNA corresponds to that of the genomic RNA. The second largest 1.6 kb and the smallest 1.3 kb dsRNAs correspond to sgRNA1 and 2 respectively.
Genome organization and replication
The genomic RNA contains four ORFs (Figure 13). However, TNV-A also contains a small 3’ proximal ORF5. ORF1 is capable of encoding an approximate 23 kDa polypeptide. Readthrough of the ORF1 amber termination codon allows translation to continue into ORF1-RT for the expression of the 82 kDa polymerase. ORF2 can encode a 7–8 kDa polypeptide implicated in cell-to-cell movement. This protein shares sequence similarity with p7-like proteins (MP1) of carmoviruses, PMV and MCMV (see Figure S3). ORF3 encodes a 6–7 kDa polypeptide that shares sequence similarity to the second MP (MP2) encoded by carmoviruses and by PMV. ORF4 encodes the 29–30 kDa CP. ORF5, present only in TNV-A, encodes a 7 kDa protein. Two sgRNAs of 1.6 and 1.3 kb are synthesized in infected cells. The smaller sgRNA is the translational template for the CP and the larger is the translational template for the ORF2 and possibly ORF3 products. Translation of TNV-D RNAs is cap and polyA-independent and is regulated by sequences in the 3′-terminal region of the genome that are similar to translational enhancer elements utilized by RCNMV but distinct from those of tombusviruses and carmoviruses. The function, if any, of an ORF5 product is not known.
Antigenic properties
Particles of necroviruses are moderately immunogenic. Species can be distinguished serologically. Antisera yield a single precipitin line in agar gel-diffusion assays.
Biological properties
Host range
Necroviruses have wide host ranges that include monocotyledonous and dicotyledonous plants. In nature, infections are typically restricted to roots. Experimental inoculations usually cause necrotic lesions on the inoculated leaves, but rarely result in systemic infection.
Transmission
Virions are readily transmitted by mechanical inoculation. Member viruses are soil-borne. Some (TNV-A, TNV-D, beet black scorch virus; BBSV) are naturally transmitted by the chytrid fungus Olpidium brassicae, while others such as olive latent virus-1 (OLV-1) are transmitted through the soil without the apparent intervention of a vector.
Geographical distribution
TNV-A and TNV-D are ubiquitous, OLV-1 was reported from several Mediterranean countries, leek white stripe virus (LWSV) from France, and BBSV from China and the US.
Cytopathic effects
Virus particles occur, often in prominent crystalline arrays, in infected cells in all tissue types, including vessels. Clumps of electron-dense amorphous material resembling accumulations of excess coat protein are present in the cytoplasm of cells infected by some necroviruses. Membranous vesicles with fibrillar material, lining the tonoplast, or derived from the endoplasmic reticulum and accumulating in the cytoplasm are elicited by LWSV or OLV-1.
Species demarcation criteria in the genus
- Extent of serological relationship as determined by immunodiffusion and/or ELISA
- Extent of sequence identity between relevant gene products
- Less than 93% aa sequence identity of the polymerase
- Less than 87% aa sequence identity of the CP
- Transmission by a fungal vector
- Natural host range
- Artificial host range reactions
List of species in the genus Necrovirus
|
Beet black scorch virus |
|
|
|
Beet black scorch virus - China |
[AF452884=NC_004452] |
(BBSV-CN) |
|
Chenopodium necrosis virus |
|
|
|
Chenopodium necrosis virus - UK |
|
(ChNV-UK) |
|
Leek white stripe virus |
|
|
|
Leek white stripe virus - France |
[X94560=NC_001822] |
(LWSV-FR) |
|
Olive latent virus 1 |
|
|
|
Olive latent virus 1 - citrus |
[X85989=NC_001721] |
(OLV-1) |
|
Olive mild mosaic virus |
|
|
|
Olive mild mosaic virus - GP |
[AY616760=NC_006939] |
(OMMV-GP) |
|
Tobacco necrosis virus A |
|
|
|
Tobacco necrosis virus A - FM-1B |
[M33002=NC_001777] |
(TNV-A-FM1B) |
|
Tobacco necrosis virus D |
|
|
|
Tobacco necrosis virus D - Hungary |
[U62546=NC_003487] |
(TNV-D-HU) |
Species names are in italic script; isolate names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Necrovirus but have not been approved as species
|
Carnation yellow stripe virus |
|
(CYSV) |
|
Furcraea necrotic streak virus |
[FJ768020] |
(FNSV) |
Genus Panicovirus
Type species Panicum mosaic virus
Distinguishing features
Virions sediment at S20,w 109S. The genomic RNA is 4.3 kb and contains four ORFs and a fifth ORF that initiates with a non-canonical GUG codon. The polymerase is larger than those encoded by other members of the family Tombusviridae. This is largely due to the presence of a region of about 20 kDa at the N-terminus of the polyprotein that is not conserved in other members of the family except the panicovirus PMV which shares a similar sequence. The virus produces a single 1.5 kb sgRNA that is a template for the expression of ORFs 2–5. The overall size and organization of the genome is similar to that of the genus Machlomovirus. However, viruses in the genus Panicovirus lack the additional 5′-proximally located ORF. The virus is restricted to monocotyledonous hosts.
Virion properties
Morphology
Virions are approximately 30 nm in diameter and exhibit icosahedral symmetry (Figure 14). Detailed capsid structure is not known. Based on CP sequence similarity, it is predicted that the capsid is structurally similar to the T=3 capsid of southern bean mosaic virus (genus Sobemovirus).
Physicochemical and physical properties
Virions sediment as one component with an S20,w of 109S, and have a buoyant density of 1.365 g cm−3 in CsCl, and a virion Mr of 6.1×106. Virus stored in desiccated tissue retained infectivity after 12 years. The thermal inactivation point of the type strain is 85 °C, and 60 °C for the St Augustine grass decline virus strain. Virions are stable at pH 6 and lower. Virions are insensitive to ether, chloroform and non-ionic detergents. Virions are stabilized by divalent cations.
Nucleic acid
Virions contain a single molecule of infectious linear positive sense ssRNA which is approximately 4.3 kb in length. The 5’ end of the RNA appears not to be capped. The RNA does not contain a 3’-terminal poly(A) tract. An approximately 1.5 kb sgRNA is also produced in vivo that appears not to be packaged into virions.
Genome organization and replication
The genomic RNA contains five ORFs (Figure 15). ORF1 encodes a 48 kDa protein. Readthrough of the ORF1 amber termination codon allows the production of a 112 kDa protein. These proteins share sequence similarity with the polymerase proteins of other members of the family (Figure S6). ORF2 encodes an 8 kDa protein that is produced by the 1.5 kb sgRNA. p8 is required for cell-to-cell movement and shows sequence similarity to the MP1 of carmoviruses, necroviruses and machlomoviruses (Figure S3) The sgRNA also encodes p6.6 from a non-canonical start codon. This protein is similar to a similarly positioned ORF for a 6.8K protein in the possible panicovirus, cocksfoot mild mosaic virus (CMMV). These proteins are similar to the MP2 proteins encoded by other members of the Tombusviridae (see Figure S4). Experimental evidence exists that p15 from ORF 4, and the CP from ORF 3 are all important for movement in addition to p8 and p6.6. PMV contains a translational enhancer in the 3′-terminal region of the genome as do several other members of the family Tombusviridae.
Antigenic properties
PMV particles are highly immunogenic. Antisera yield a single precipitin line in agar gel-diffusion assays. There are several serological strains of the type strain as well as the serologically distinct St Augustine grass decline virus strain.
Biological properties
Host range
In nature, PMV is restricted to grass species in the Paniceae tribe of the Poaceae. It is known to cause diseases of note in switch grass (Panicum virgatum), St Augustine grass (Stenotaphrum secundatum) and centipede grass (Eremochloa ophiuroides). In the laboratory, a number of additional species in the Graminae can be symptomless hosts. Zea mays is used as a propagation host for the type strain. St Augustine grass must be used as a propagation host for the St Augustine grass decline virus strain. CMMV infects grass species such as Dactylis glomerata (cocksfoot), Phleum pratense (Timothy), Holcus sp., Bromus sp. and Festuca sp.
Transmission
The virus is readily transmitted by mechanical inoculation. The virus is typically transmitted by the transport and replanting of infected sod. There is one report that the St Augustine grass decline virus strain was seed-transmitted through Setaria italica.
Geographic distribution
PMV has been reported in the US and Mexico. The type strain is widely distributed in a number of turf grasses throughout the central United States whereas the St Augustine grass decline virus strain is widely distributed throughout the southern US. CMMV is present in Europe and Canada.
Pathogenicity, association with disease
The virus typically forms a systemic mosaic. More severe symptoms, including chlorotic mottling, stunting and seed yield reduction, occur in forage grasses when PMV is in a mixed infection with panicum mosaic satellite virus. The St Augustine grass decline virus strain can cause a severe disease on St Augustine grass with symptoms being more severe in the hot summer months.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Panicovirus
|
Panicum mosaic virus |
|
|
|
Panicum mosaic virus - Kansas |
[U55002=NC_002598] |
(PMV-KAN) |
Species names are in italic script; isolate names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Panicovirus but have not been approved as species
|
Cocksfoot mild mosaic virus |
[EU081018=NC_011108] |
(CMMV) |
|
Molinia streak virus |
|
(MoSV) |
Genus Machlomovirus
Type species Maize chlorotic mottle virus
Distinguishing features
Virions sediment at an S20,w of 109S. The genomic RNA is about 4.4 kb and contains four ORFs. Like that of panicoviruses, the polymerase is larger, containing an N-terminal extension of about 20 kDa that is not present in the polymerases of other members of the family. Unlike other family members, the ORF for the polymerase is not the 5′-most proximal ORF. A 1.47 kb sgRNA is present in infected plants. Apparently a 0.34 kb sgRNA is present in infected plants that does not contain an ORF for any viral protein. The overall size and organization of the genome is quite similar to that of the genus Panicovirus. However, genomes of machlomoviruses encode an additional 5′-proximally located ORF encoding a 32 kDa protein of unknown function. The virus is restricted to monocotyledonous hosts.
Virion properties
Morphology
Virions are approximately 30 nm in diameter and exhibit icosahedral symmetry (Figure 16). Detailed structure of virions is not known. Based on CP sequence similarity, it is predicted that the capsid is structurally similar to the T=3 capsids of sobemoviruses.
Physicochemical and physical properties
Mr of virions is 6.1×106; S20,w is 109S; buoyant density in CsCl is 1.365 g cm−3. Virions are insensitive to ether, chloroform and non-ionic detergents. Virions are stable in vitro for up to 33 days and the thermal inactivation point of virions is between 80–85 °C. Virions are stable at pH 6 and lower. Virions are stabilized by divalent cations.
Nucleic acid
Virions contain a single molecule of infectious linear positive sense ssRNA. The RNA is 4437 nt in length. The 5’ end of the RNA is probably uncapped The RNA does not contain a 3’-terminal poly(A) tract. Either a 1470 or an 1100 nt sgRNA is also packaged into virions at a very low level.
Genome organization and replication
The genomic RNA contains four ORFs (Figure 17). ORF1 is capable of encoding a 32 kDa protein. ORF2 can encode a 48 kDa protein. Readthrough of the ORF2 amber termination codon allows for translation to continue into ORF2-RT, yielding a 112 kDa protein which shares sequence similarities with polymerase proteins of other members of the family (see Figure S6). ORF3 encodes a 7 kDa protein whose carboxyl- terminus is like those of the MP1 proteins encoded by the similarly located small ORFs in the genomes of carmoviruses, necroviruses and panicoviruses (see Figure S3). A protein corresponding to MP2 does not appear to be encoded by the MCMV genome. If the ORF3 opal termination codon is read through, a 33 kDa protein would be produced. ORF4 encodes the 25 kDa CP and appears to be translated from the 1.47 kb sgRNA. A second 0.34 kb sgRNA is made in vivo; however, it is not known if it acts as a mRNA. The functions of proteins encoded by ORF1 and the ORF3 readthrough product are not known.
Antigenic properties
MCMV particles are moderately to highly immunogenic. Serological variants have been identified. Antisera yield a single precipitin line in agar gel-diffusion assays.
Biological properties
Host range
In nature, the virus systemically infects varieties of maize (Zea mays). In the laboratory, the virus is restricted to members of the family Gramineae.
Transmission
The virus is readily transmitted by mechanical inoculation. The virus is also seed-transmitted. Kansas and Nebraska isolates can be transmitted by six species of chrysomelid beetles in the laboratory. A Hawaiian isolate is transmitted by thrips.
Geographic distribution
The virus has been reported in Argentina, Mexico, Peru and the United States. Within the United States, the virus is restricted to the Republican River valley of Kansas and Nebraska, and to Kauai, Hawaii.
Pathogenicity, association with disease
MCMV causes a mild mosaic on maize in nature. When plants are also infected with one of several Graminae-specific potyviruses, a severe necrotic disease results, termed corn lethal necrosis.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Machlomovirus
|
Maize chlorotic mottle virus |
|
|
|
Maize chlorotic mottle virus - United States |
[X14736=NC_003627] |
(MCMV-US) |
Species names are in italic script; isolate names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Machlomovirus but have not been approved as species
None reported.
List of unassigned species in the family Tombusviridae
|
Maize necrotic streak virus |
|
|
|
Maize necrotic streak virus – Arizona |
[AF266518=NC_007729] |
(MNeSV-AZ) |
|
Pelargonium line pattern virus |
|
|
|
Pelargonium line pattern virus - PV-0193 |
[AY613852=NC_007017] |
(PLPV-PV0193) |
Species names are in italic script; isolate names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
Phylogenetic relationships within the family
The polymerase is highly conserved among members of the family Tombusviridae with the readthrough (or frameshift) portion showing the greatest level of conservation, i.e. 27–60% identity among type members compared to 2–20% identity in the pre-readthrough (pre-frameshift) region. Phylogenetic analyses (see Figure S6) show that overall the polymerases of members of a genus are more closely related to each other than they are to members of other genera consistent with the notion that such trees are a useful criterion for defining genera. An exception to this is the polymerase of the carmovirus galingsoga mosaic virus which appears to be more closely related to necroviruses. Also, necroviruses are divided into two major groupings: one that is associated with the tombusvirus/aureusvirus lineage and the other with the carmovirus/machlomovirus/panicovirus lineage.
Sequence comparisons of the MPs indicate that there are three main groupings that are phylogenetically unrelated. The dianthoviruses encode a 32 kDa MP which is not related to the MPs of other members of the family. The tombusviruses and aureusviruses share a similar movement protein (Figure 1). The remaining viruses with the exception of the machlomoviruses, encode two MPs that fall into two classes, MP1 and MP2 (see Figure S3,4). Machlomoviruses encode an MP1-like protein but not an MP2-like protein. Phylogenetic analysis of the MP2-like protein may be a useful taxonomic tool since members of a genus show a greater phylogenetic relationship to each other than they do to members of other genera (see Figure S4). MP1 comparisons would not appear to be useful since the phylogenetic groupings are weak and are not in agreement with current taxonomic groupings.
The CPs are conserved among all genera and overall the CP is a reliable indicator of taxonomic identity (see Figure S5). However, exceptions exist. The CPs of tombusviruses and aureusviruses do not separate into distinct groupings. Also, sometimes the CP of a virus does not group with other members of its genus. Examples include the CPs of the carmoviruses melon necrotic spot virus (MNSV) and pea stem necrosis virus (PSNV) that are present in a distinct lineage. This is also the case for the aureusvirus CLSV which does not group with the tombusvirus/aureusvirus lineage. On a structural level, the CPs fall into two main classes: those with a protruding domain (dianthoviruses, tombusviruses, aureusviruses, avenaviruses and carmoviruses) and those without a protruding domain (necroviruses, machlomoviruses and panicoviruses).
Similarity with other taxa
The polymerases of members of the family Tombusviridae are related to the polymerases of the genus Luteovirus, as exemplified by barley yellow dwarf virus-PAV, as well as the genus Umbravirus as exemplified by pea enation mosaic virus-2 (see Figure 18; Figure S5). The luteovirus polymerase is most closely related to that of the dianthoviruses. The polymerases are also distantly related to those of the genera Hepacivirus and Pestivirus in the family Flaviviridae. The CPs of necroviruses are phylogenetically related to the CPs of sobemoviruses (Figure 18; Figure S5). The hepatitis E virus and the betanodavirus CPs are distantly related to those of members of the family Tombusviridae (Figure 18). The dianthovirus MP is phylogenetically related to the movement proteins of a diverse number of viruses including members of the families Bromoviridae, Rhabdoviridae, Bunyaviridae, Virgaviridae and the genus Umbravirus. It has recently been found that the MPs of tombusviruses and aureusviruses share significant sequence similarity with the MPs of members of the genus Ourmiavirus (see Figure S7).
Derivation of names
Aureus: from the specific epithet of Scindapsus aureus (pothos), natural host of the virus.
Avena: from Avena, the generic name for oats.
Carmo: from carnation mottle.
Diantho: from Dianthus, the generic name of carnation.
Machlomo: from maize chlorotic mottle.
Necro: from Greek nekros, “dead body”.
Panico: from panicum mosaic.
Tombus: from tomato bushy stunt.
Supplementary Figures
Supplementary Figures, Figure S1 to S7, are available in the right-hand menu above.
Further reading
Giesman-Cookmeyer et al., 1995 D. Giesman-Cookmeyer, K.H. Kim, S.A. Lommel, Dianthoviruses. In: . Pergamon Press, Oxford1995157–176.
Hamilton and Tremaine, 1996 R.I. Hamilton, J.H. Tremaine, Dianthoviruses: properties, molecular biology, ecology, and control. In: . Plenum Press, New York1996251–282.
Koenig, 1988 R. Koenig, R. Koenig, Polyhedral plant viruses with monopartite RNA genomes. Introduction and summary of important propertiesThe Plant Viruses: Polyhedral Virions with Monopartite RNA Genomes. In: R. Koenig, The Plant Viruses: Polyhedral Virions with Monopartite RNA Genomes. Plenum Press, New York and London1988.
Koenig et al., 2004 R. Koenig, J.T. Verhoeven, C.E. Fribourg, E. Pfeilstetter, D.E. Lesemann, Evaluation of various species demarcation criteria in attempts to classify ten new tombusvirus isolates. Arch. Virol. 149 (2004) 1733–1744.
Miller et al., 2007 W.A. Miller, Z. Wang, K. Treder, The amazing diversity of cap-independent translation elements in the 3’-untranslated regions of plant viral RNAs. Biochem. Soc. Trans. 35 (2007) 1629–1633.
Morris and Carrington, 1988 T.J. Morris, J.C. Carrington, R. Koenig, Carnation mottle virus and viruses with similar propertiesThe Plant Viruses: Polyhedral Virions with Monopartite RNA Genomes. In: R. Koenig, The Plant Viruses: Polyhedral Virions with Monopartite RNA Genomes. New York and London, Plenum Press198873–112.
Rastgou et al., 2009 M. Rastgou, M.K. Habibi, K. Izadpanah, V. Masenga, R.G. Milne, Y.I. Wolf, E.V. Koonin, M. Turina, Molecular characterization of the plant virus genus Ourmiavirus and evidence of inter-kingdom reassortment of viral genome segments as its possible route of origin. J. Gen. Virol. 90 (2009) 2525–2535.
Russo et al., 1994 M. Russo, J. Burgyan, G.P. Martelli, Molecular biology of Tombusviridae. Adv. Virus Res. 44 (1994) 381–428.
White and Nagy, 2004 K.A. White, P.D. Nagy, Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucl. Acid Res. Mol. Biol. 78 (2004) 187–226.
Contributed by
Rochon, D., Lommel, S., Martelli, G.P., Rubino, L. and Russo, M.
Figures
Figure 1 (Top row, left) Computer reconstruction of a tomato bushy stunt virus (TBSV) particle based on X-ray crystallography at 2.9 resolution (J.Y. Sgro, University of Wisconsin-Madison) (Olson et al. (1983). J. Mol. Biol., 171, 61-93). (Top row center) Diagrammatic representation of a T=3 TBSV particle. A, B and C correspond to the three conformational states of the CP. (Top row, right) Negative contrast electron micrograph of TBSV particles. (Bottom row, left) Computer reconstruction of a tobacco necrosis virus A (TNV-A) particle, based on X-ray crystallography at 2.25 resolution (Oda et al. (2000). J. Mol. Biol., 300, 153-169). (Bottom row center) Schematic representation of the T=3 structure of TNV-A particles. A, B, C correspond to the three conformational states of the CP. (Bottom row, right) Negative contrast electron micrograph of particles of TNV (species unidentified). The bars represent 50 nm.
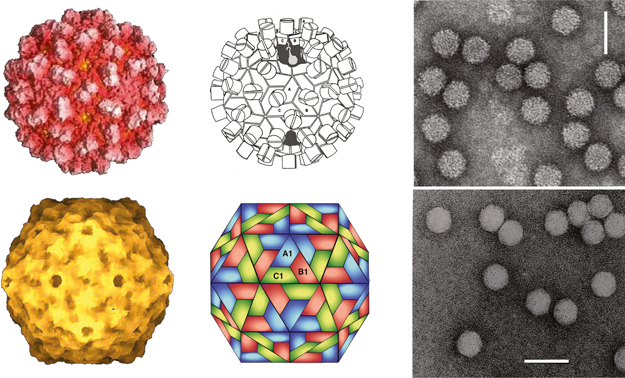
Figure 2 Genome organization of the type species for each genus in the family Tombusviridae. Boxes represent known and predicted ORFs. Similarly shaded boxes represent proteins with sequence conservation with the exception of the gray boxes which represent proteins unique to the indicated virus. The yellow boxes represent ORFs encoding the phylogenetically conserved polymerase. Red-hatched boxes represent CP encoding ORFs. Right-hatched boxes identify CPs lacking a protruding domain that are related to those of the genus Sobemovirus while those which are left-hatched represent tombusvirus-like CPs that contain protruding domains. Blue boxes represent viral MPs. In carmoviruses, necroviruses and panicoviruses the light-blue-shaded box corresponds to a second movement protein conserved among these viruses. In avenaviruses the dark-blue-shaded box corresponds to a potential movement protein that does not show sequence similarity to any of the other movement proteins encoded by the Tombusviridae. In tombusviruses and aureusviruses the green-shaded boxes correspond to the silencing suppressor protein. The orange-shaded region in PMV and MCMV are similar to each other but do not have sequence similarity with other members of the Tombusviridae. RT: translational readthrough of termination codon. 1FS: 1 ribosomal frameshifting event. CP: capsid protein.
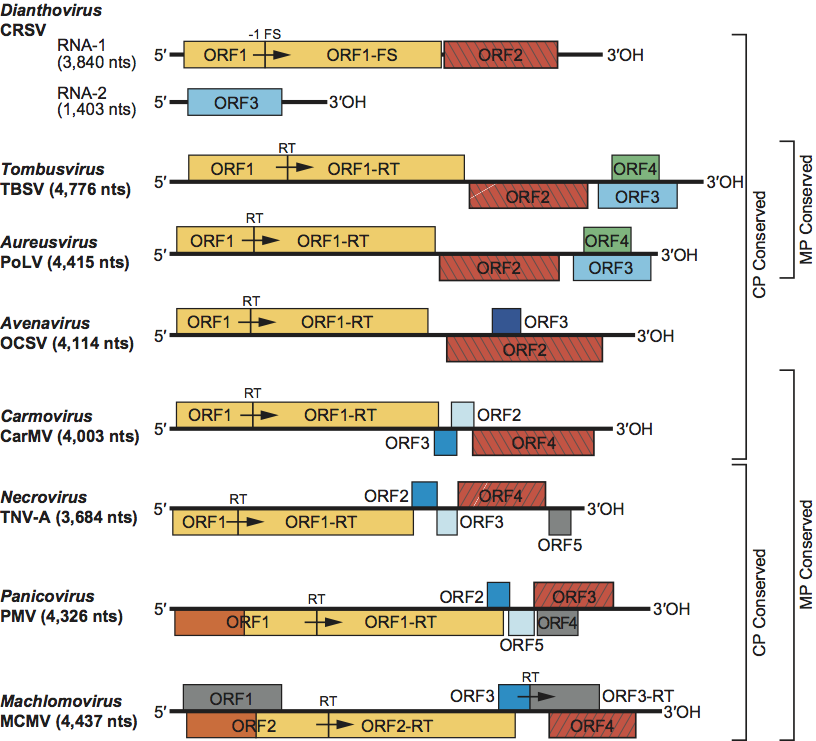
Figure 3 (Left) Cryo reconstruction of the CNV particle showing the P-domain pairs and the five-fold, three-fold and two-fold axes of symmetry. (Center) Cut away image of CNV particle showing features of the structured interior including the inner shell and the connections between it and the outer shell at the particle three-fold axes (cryo images kindly provided by Tom Smith, Donald Danforth Plant Science Centre, St Louis, MO). (Right) Negative contrast electron micrograph of TBSV particles. The bar represents 50 nm.
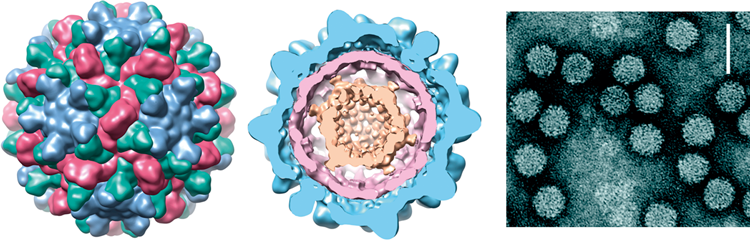
Figure 4 Genome organization and replication strategy of the tombusvirus tomato bushy stunt virus (TBSV). Boxes represent known and predicted ORFs with the sizes of the respective proteins (or readthrough products) shown on the horizontal bars below the ORFs. The yellow-shaded ORFs indicate polymerase proteins that have a high degree of sequence conservation within the family Tombusviridae. Left-hatched box identifies the CP that is conserved among other genera within the family Tombusviridae that have a protruding domain. The blue box shows the MP which shares sequence similarity to the MPs of aureusviruses (see Figure S1). The green box identifies the ORF encoding the silencing suppressor that shares sequence similarity with that of aureusviruses (see Figure S1). Lines underneath depict the two sgRNAs that are synthesized in infected cells. The bars underneath the sgRNAs correspond to the encoded proteins, i.e., the CP, the RNA silencing suppressor and the movement protein (MP). RT=amber termination codon that is read through.
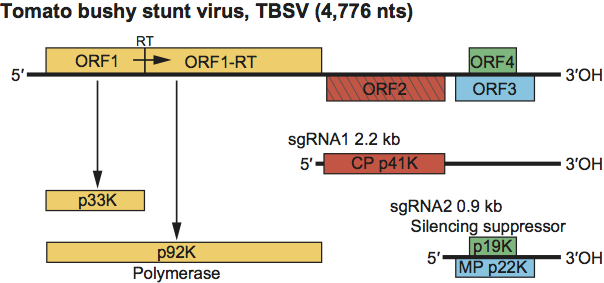
Figure 5 Cryo-EM of the RCNMV particle (from Baker, Sherman and Lommel, with permission; see also Sherman, M.B. et al. (2006). J. Virol., 80, 10395-10406). The RCNMV virion has an ordered internal shell as can be seen by the cross-section of the cryo structure (center panel). In addition to an outer shell (left panel, colored yellow) comprised entirely of coat protein, RCNMV has an additional internal ordered density (colored with shades of blue). Approximately 90% of RCNMVs density corresponds to the coat protein. Thus it is possible that the ordered density observed in the inner cage may correspond in part to ordered RNA.
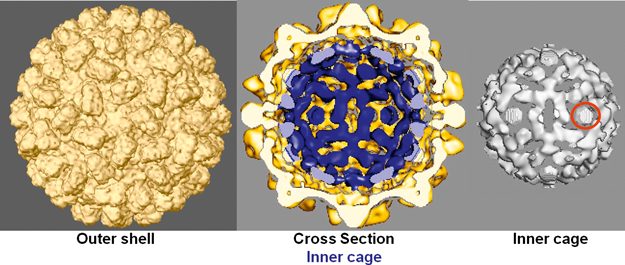
Figure 6 Genome organization and replication strategy of the dianthovirus carnation ringspot virus (CRSV). Boxes represent known ORFs with the sizes of the respective proteins (or readthrough products) indicated in the bars below. Yellow ORFs on RNA1 indicate polymerase proteins that have a high degree of sequence conservation within the family Tombusviridae. Left-hatched box on RNA1 identifies the CP that is conserved among other genera within the family Tombusviridae that have a protruding domain. The blue box on RNA2 identifies the ORF that encodes the MP which is also a silencing suppressor. The MP contains a region of sequence similarity with MPs in the other virus families (see text). The line under RNA1 depicts the 1.5 kb CP sgRNA. 1 FS=site of 1 ribosomal frameshifting.
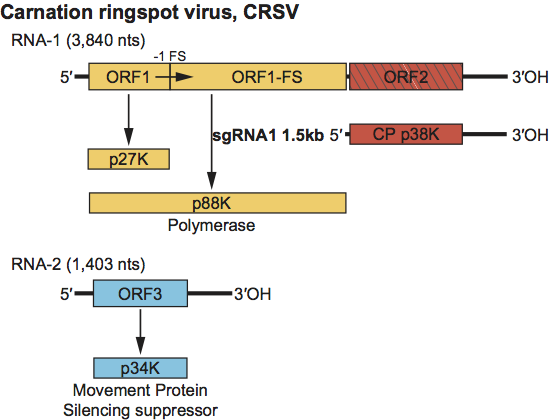
Figure 7 (Left) Diagrammatic representation of a T=3 particle with A, B, C corresponding to the three conformational states of the CP subunit. (Right) Negative contrast electron micrograph of pothos latent virus particles. The bar represents 50 nm.
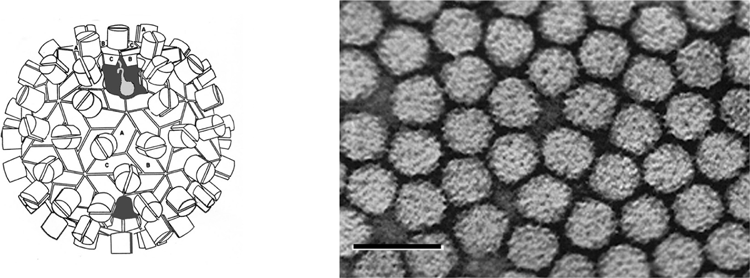
Figure 8 Genome organization and replication strategy of PoLV. Boxes represent known and predicted ORFs with the sizes of the respective proteins (or readthrough products) indicated in the bars below. Yellow ORFs indicate polymerase proteins that have a high degree of sequence conservation within the family Tombusviridae. Left-hatched box identifies the CP that is highly conserved among other genera within the family Tombusviridae that share a protruding domain. The blue box corresponds to the MP which shows sequence similarity to the MP of tombusviruses (see Figure S1). The green box identifies the silencing suppressor that shares sequence similarity with that of tombusviruses (see Figure S1). Lines underneath depict the two sgRNAs that are synthesized in infected cells and which allow for expression of the CP, p14K and the MP. RT=termination codon that is read through.
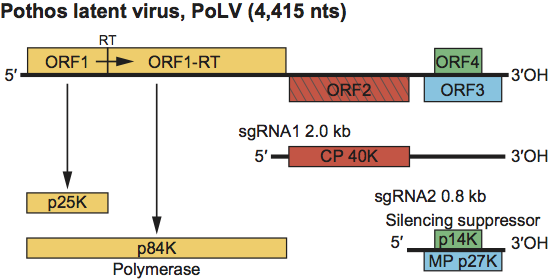
Figure 9 Genome organization of the avenavirus oat chlorotic stunt virus (OCSV). Boxes represent known and predicted ORFs with the sizes of the respective proteins (or readthrough products) indicated in the bars below. Yellow ORFs indicate polymerase proteins that have a high degree of sequence conservation within the family Tombusviridae. Left-hatched box identifies the CP that is highly conserved among other genera within the family Tombusviridae that share a protruding domain. The blue box identifies a putative cell-to-cell MP; this protein does not share sequence similarity with any of the other Tombusviridae MPs. RT=termination codon that is read through.
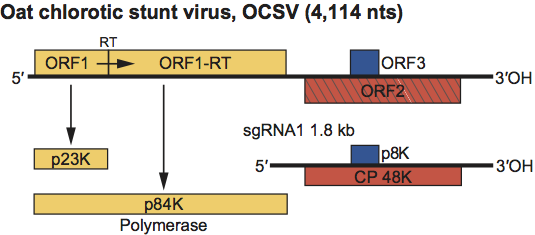
Figure 10 (Left) Computer reconstruction of a carnation mottle virus (CarMV) particle based on X-ray crystallography at 3.2 resolution (from Morgunova et al. (1994). FEBS Letters, 338, 267-271; with permission). (Center) Diagrammatic representation of a carmovirus particle. (Right) Negative contrast electron micrograph of CarMV particles. The bar represents 50 nm.
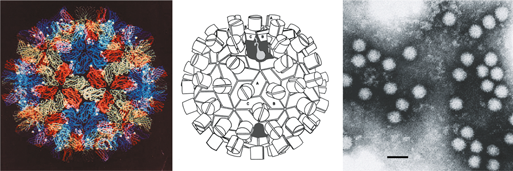
Figure 11 Genome organization and replication strategy of the carmovirus CarMV. Boxes represent known and predicted ORFs with the sizes of the respective proteins (or readthrough products) indicated within the horizontal bars below. Yellow ORFs indicate polymerase proteins that have a high degree of sequence conservation within the family Tombusviridae. Left-hatched box identifies the CP that is conserved among other genera in the family Tombusviridae that have a protruding domain. The dark blue box identifies one (MP1) of the two proteins involved in cell-to-cell movement that exhibits carboxyl-terminal sequence conservation with similar proteins in the machlomoviruses, necroviruses and panicoviruses (see Figure S2). The light-blue box identifies an ORF for a second movement protein (MP2), that shares sequence similarity with the small proteins encoded by viruses in the genera Necrovirus and Panicovirus (see Figure S3). Lines underneath the gene map depict the two sgRNAs that are synthesized in infected cells that encode MP1, MP2 and CP. RT=amber termination codon that is read through. The CP of CarMV is indicated as a suppressor of silencing by analogy to the suppressor activity of other carmovirus CPs (see text).
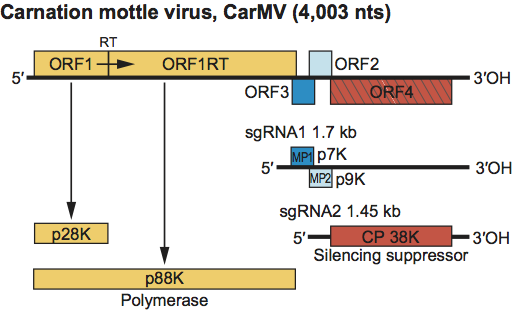
Figure 12 (Left) Computer reconstruction of a tobacco necrosis virus A (TNV-A) particle based on X-ray crystallography at 2.25 resolution (Oda et al. (2000). J. Mol. Biol., 300, 153-169). (Center) Diagrammatic representation of a necrovirus particle. VP1, VP2 and VP3 correspond to the three conformational states of the CP subunit. (Right) Negative contrast electron micrograph of TNV-A virions. The bar represents 50 nm.
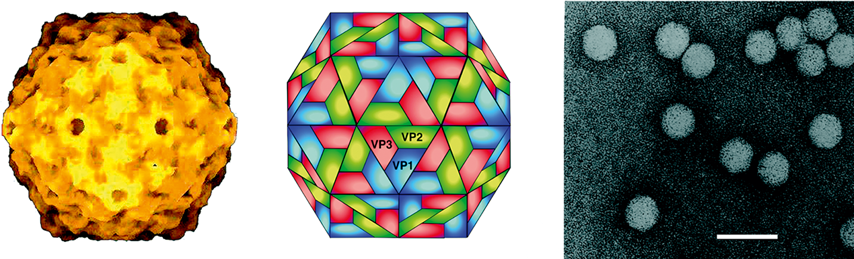
Figure 13 Genome organization and replication strategy of the necrovirus TNV-A. Boxes represent known and predicted ORFs with the sizes of the respective proteins (or readthrough products) indicated in the bars below. Shaded ORFs indicate polymerase proteins that have a high degree of sequence conservation within the family Tombusviridae. Right-hatched box identifies the CP that is highly conserved among other genera within the family Tombusviridae that lack a protruding domain. The dark-blue box identifies one (MP1) of the two proteins involved in cell-to-cell movement that exhibits carboxyl-terminal sequence conservation with similar proteins in the machlomoviruses, carmoviruses, and panicoviruses (see Figure S2). The light-blue box identifies an ORF for a second movement protein (MP2), that shares sequence similarity with the small proteins encoded by viruses in the genera Carmovirus and Panicovirus (see Figure S3). The ORF5 protein does not share sequence similarity with other viral proteins in the family Tombusviridae. Lines underneath the gene map depict the two sgRNAs that are synthesized in infected cells and which allow for the expression of MP1, MP2 and CP. RT=amber termination codon that can be read through.
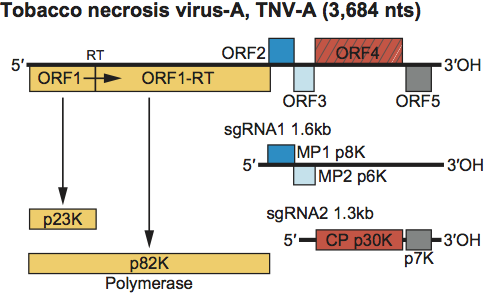
Figure 14 (Left) Diagrammatic representation of a particle of panicum mosaic virus (PMV). A, B and C correspond to the three conformational states of the CP subunit. (Right) Negative contrast electron micrograph of PMV particles. The bar represents 100 nm.
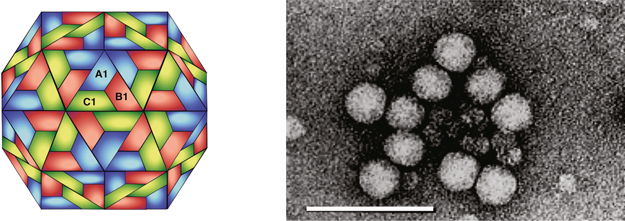
Figure 15 Genome organization and replication strategy of the panicovirus PMV. Boxes represent known and predicted ORFs with the sizes of the respective proteins (or readthrough products) indicated in the bars below. Shaded ORFs indicate polymerase proteins that have a high degree of sequence conservation within the family Tombusviridae. Right-hatched box identifies the CP that is highly conserved among other genera within the family Tombusviridae that lack a protruding domain. The dark-blue box identifies one (MP1) of the two proteins involved in cell-to-cell movement that exhibits carboxyl-terminal sequence conservation with like proteins in the machlomoviruses, necroviruses, and carmoviruses (see Figure S2). The light-blue box identifies an ORF for a second movement protein (MP2), that shares sequence similarity with the small proteins encoded by viruses in the genera Necrovirus and Carmovirus (see Figure S3). The 1.5 kb sgRNA is illustrated as a line below the genomic RNA and is believed to encode the four indicated proteins. RT=amber termination codon that can be read through. 1 FS=Site of a 1 ribosomal frameshift event.
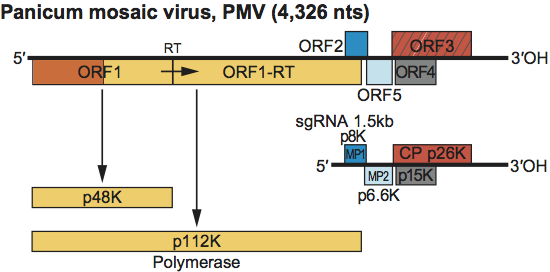
Figure 16 (Left) Diagrammatic representation of a particle of MCMV. VP1, VP2 and VP3 correspond to the three conformational states of the CP subunit. (Right) Negative contrast electron micrograph of MCMV virions. The bar represents 100 nm.
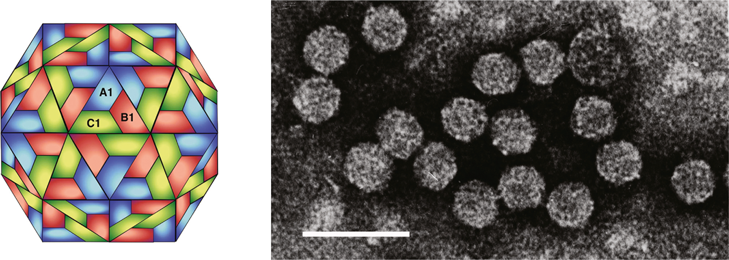
Figure 17 Genome organization and replication strategy of the machlomovirus MCMV. Boxes represent known and predicted ORFs with the sizes of the respective proteins (or readthrough products) indicated in the bars below. Shaded ORFs indicate polymerase proteins that have a high degree of sequence conservation within the family Tombusviridae. Right-hatched box identifies the CP that is highly conserved among those genera within the family Tombusviridae that lack a protruding domain. The blue box identifies the putative cell-to-cell MP that exhibits sequence conservation with similar proteins in the carmoviruses, necroviruses and panicoviruses (see Figure S2). The gray boxes identify ORFs having no significant sequence similarity with other viral proteins. The 1.47 kb sgRNA with its encoded proteins as well as the 0.34 kb sgRNA are illustrated as lines below the genomic RNA. RT=termination codon that is read through.
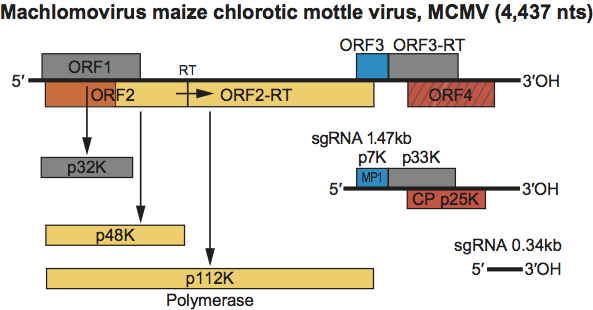
Figure 18 Phylogenetic analysis of CP and polymerase proteins of genera of the family Tombusviridae and relationships to other taxa. The coloured boxes encompass viruses with the two different morphological types of virions, smooth (purple) versus those with a protruding domain (yellow). Protein sequences were aligned and dendograms were constructed using ClustalX2. Genera of the family Tombusviridae are in bold whereas non-members are in regular type. The CP of plum pox virus (family Potiviridae) was used as an outgroup. Barley yellow dwarf virus-PAV is a luteovirus (family Luteoviridae) and southern bean mosaic virus (SBMV) is a sobemovirus. Striped jack nervous necrosis virus (SJNNV) is the type member of the genus Betanodavirus in the family Nodaviridae. A more extensive dendogram that includes the CPs of all members of the Tombusviridae is shown in Figure S5. Polymerase dendogram. Hepatitis virus C-1 (HCV-1) and bovine viral diarrhea virus (BVDV-1) are members of the genera Hepacivirus and Pestivirus, respectively in the family Flaviviridae. Pea enation mosaic virus-2 (PEMV-2) is an umbravirus in the family Luteoviridae and Hepatitis E virus is a hepevirus in the family Hepeviridae. The polymerase of tobacco mosaic virus (family Virgaviridae) was used as an outgroup for generation of the polymerase dendogram. A dendogram depicting relationships of the polymerases among all members of the Tombusviridae is shown in Figure S6.
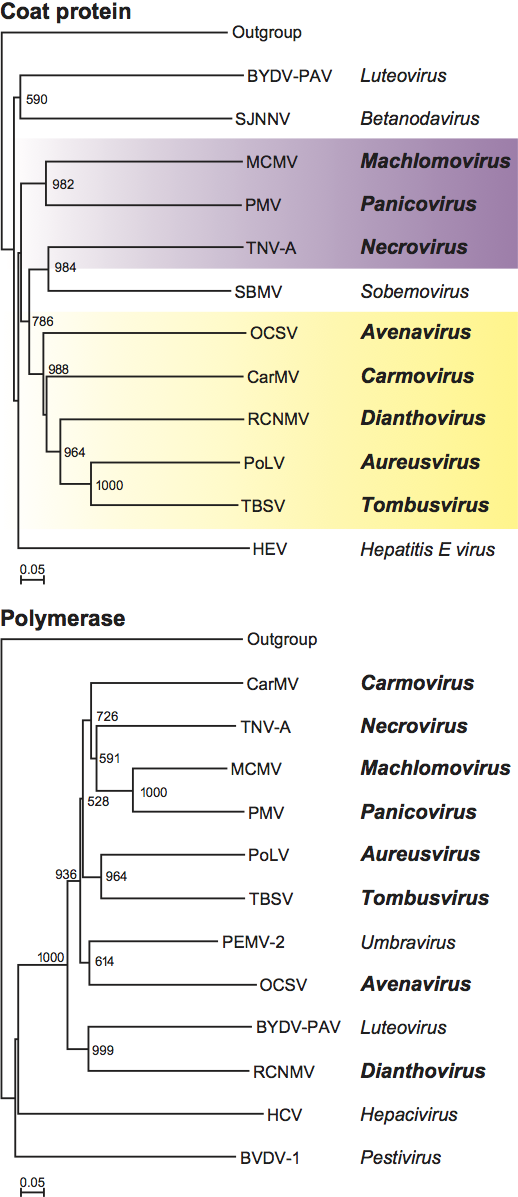
Figure S1.
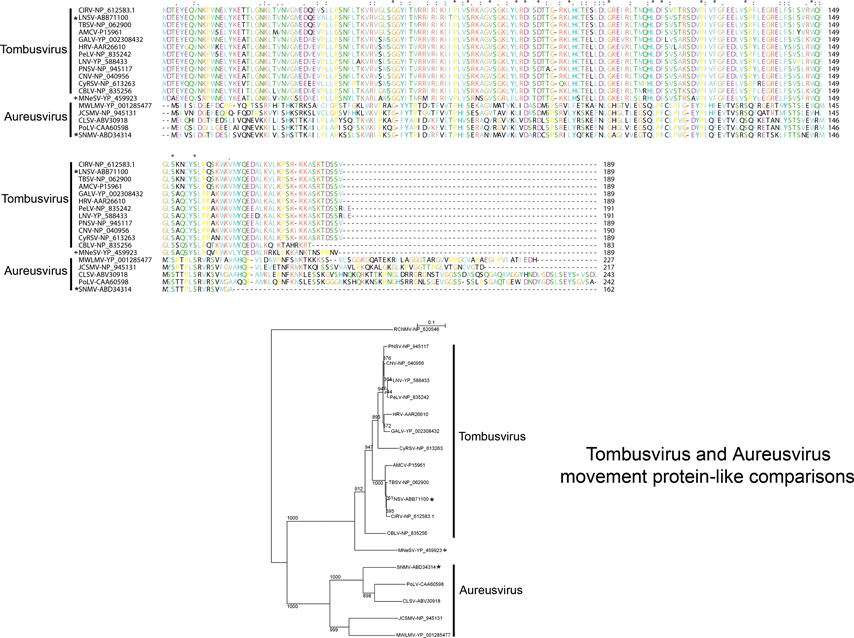
Figure S2.
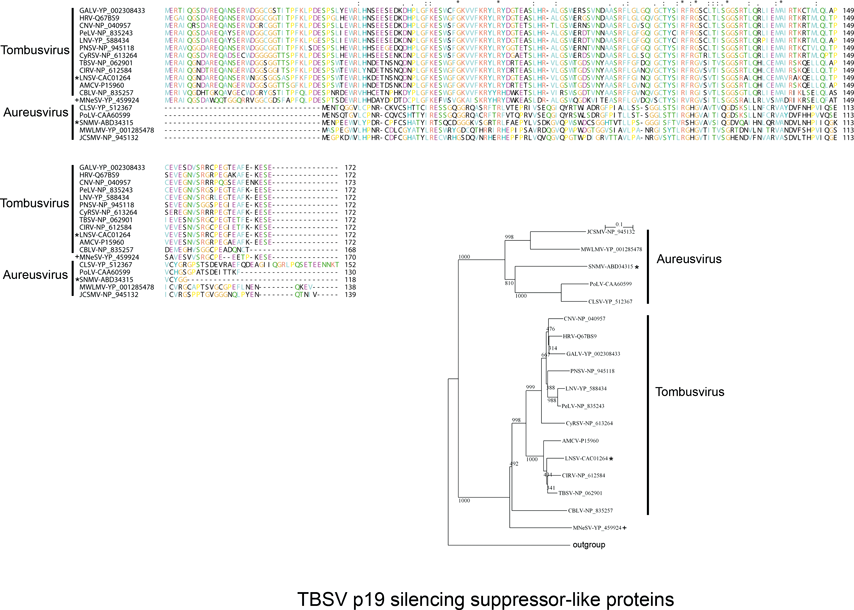
Figure S3.
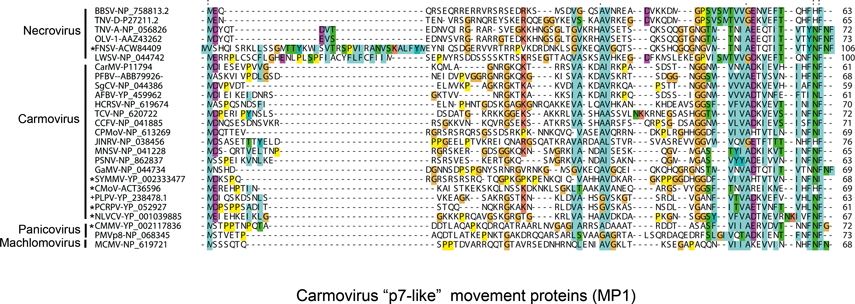
Figure S4.
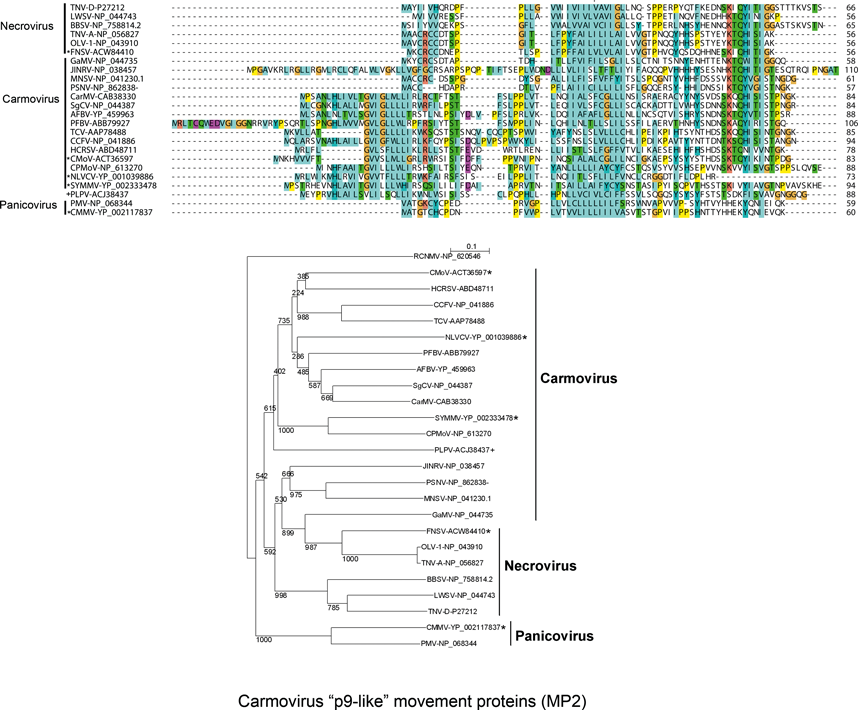
Figure S5.

Figure S6.
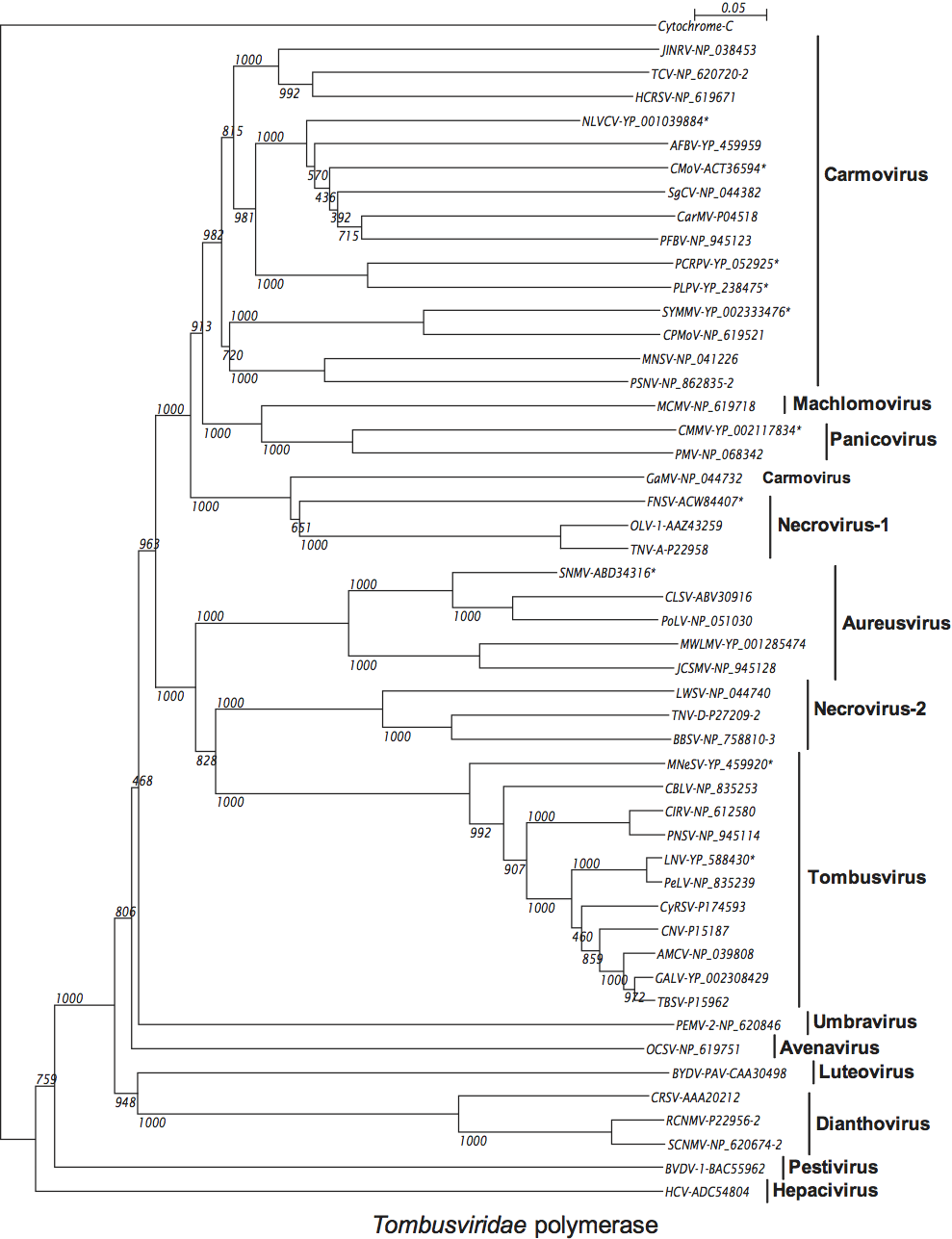
Figure S7.

