Family: Togaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Alphavirus virions are assembled into icosahedral particles of approximately 70 nm in diameter and a molecular mass of 5.2×107 daltons. The particle core (40 nm) consists of 240 copies of the capsid protein (C or CP) arranged in a T4 symmetry surrounding the genomic RNA. However, in rubiviruses (Rubella virus, RUBV), the core particle consists of multiple CP disulfide-linked homodimers with the genomic RNA. The nucleocapsid core is covered by a lipid bilayer containing heterodimers of the two virally encoded glycoproteins, E2 and E1, which form a regular icosahedral surface lattice. The E1/E2 heterodimers of Sindbis virus (SINV) have been shown to be capable of producing three distinct morphological structures of varying dimensions. These heterodimers undergo conformational changes during the process of cellular entry. The lipid bilayer is derived from the cellular plasma membrane. RUBV virions are just over 60 nm in diameter and exhibit a T3 symmetry rather than a T4 as seen with the alphaviruses. Structural analysis of RUBV has been limited by an inability to purify virions free of cell-derived membranes. (See Figure 1.)
Physicochemical and physical properties
Alphaviruses have a buoyant density in sucrose gradients of 1.20 g cm−3 while that of RUBV is slightly less at 1.18 to 1.19 g cm−3. The sedimentation coefficient of togavirus virions ranges from 240S to 350S with most of the variability associated with RUBV (Alphaviruses have an S20,w of 280S). Denaturing agents including formaldehyde and beta-propiolactone, detergents, acid, and lipid solvents readily eliminate infectivity. Togavirus infectivity is also abrogated by heat inactivation with virions being fairly heat labile; treatment at 58 °C results in a half-life of minutes. Exposure to ultravirolet light or radiation also results in loss of infectivity.
Nucleic acid
All togaviruses contain a single strand of positive sense RNA as their genetic material. The size ranges from 9.7 to 11.8 kb for the alphaviruses and is approximately 9.8–10.0 kb for RUBV. The RNA contains a 5′-terminal m7G cap and a 3′-terminal poly-A tail.
Proteins
There are four non-structural proteins, designated nsP1-4, which encode the viral replication machinery of the togaviruses. These proteins are not found in intact virions but only in infected cells. The structural proteins consist of the capsid protein (C or CP), the two surface envelope glycoproteins (E1 and E2) and two small peptides (E3 and 6K) which serve as leader peptides for E2 and E1 respectively but are not present in all alphaviruses. These peptides are not present in the rubiviruses.
Lipids
Virion associated lipids are derived from the host cell membranes during the process of virus maturation and budding. Budding occurs at the plasma membrane only for the alphaviruses while RUBV buds from both intracellular and surface membranes. These lipids make up approximately 30% of the total weight of the virion and their composition varies with the cell from which the virions were derived. When alphaviruses replicate in mammalian cells, particles have significantly more cholesterol than when the virus is propagated in invertebrate cells.
Carbohydrates
Both E1 and E2 glycoproteins of alphaviruses contain N-linked glycans while O-linked glycans are present only on the RUBV E2. Alteration of the glycosylation can negatively affect viral replication. In RUBV, carbohydrates make up 10% of the mass of E1 and 30–40% of E2.
Genome organization and replication
The RNA genome encodes the non-structural proteins in a single ORF immediately after a 5′ non-coding region. The proteins are oriented as 5′ nsP1 (methyltransferase and guanyltransferase) – nsP2 (helicase and protease) – nsP3 (phosphoprotein integral in minus strand synthesis) – nsP4 (RNA dependent RNA polymerase) – 3′. There is a stop codon present between the nsP3 and nsP4 genes in the majority of alphaviruses resulting in a limited amount of nsP1234 generated by inefficient read-through. The non-structural genes occupy approximately two-thirds of the genome. Immediately downstream of the non-structural ORF, there is a promoter for transcription of the subgenomic mRNA from which the structural polyprotein is translated. The structural proteins include the capsid (C/CP), E3, E2, 6K, and E1 proteins. The structural ORF is followed by a non-coding region of varying length (range 77–609 nucleotides) and finally, a polyadenylation signal. (See Figure 2.)
While RUBV and the alphaviruses share homology in the cis-acting elements at the 5′ end of the genome and in the subgenomic promoter region, there are major differences in their replication processes. In alphaviruses, the initial polyproteins generated, P123 and/or P1234, are cleaved in trans by the viral protease activity of the nsP2 protein. The P123 precursor protein combined with nsP4 form the replication complex associated with minus-strand replication. This negative-strand serves as the template in the synthesis of a full-length, positive sense RNA that will eventually be encapsidated as well as a subgenomic 26S mRNA that encodes the viral structural proteins. Cleavage of the P123 polyprotein generates nsP1, nsP2 and nsP3 individual proteins which are involved in positive-strand synthesis. In RUBV, the polyprotein precursor is cleaved either in cis or trans into two products, P150 and P90, by a protease located near the C-terminus of P150. The order of functional motifs in the RUBV genome is different from that of the alphaviruses with P90 polyprotein containing both helicase and replicase motifs.
The alphavirus subgenomic mRNA is translated into a single polyprotein from which individual structural proteins are produced by both viral and cellular proteases. RUBV CP lacks the viral autoprotease activity of the alphavirus C protein and thus relies completely upon cellular endopeptidases for structural protein cleavage. The glycoproteins that are produced are inserted into the endoplasmic reticulum during translation and are translocated to the plasma membrane for alphaviruses or to intracellular membranes for RUBV. Upon generation of sufficient C protein, this protein assembles with the viral RNA to form the viral nucleocapsids. This process occurs in the cytosol for alphaviruses and during the budding process associated with the Golgi apparatus for RUBV. Budding through the plasma membrane (alphaviruses) or the Golgi and plasma membrane (RUBV) leads to the acquisition of a lipid envelope containing the two membrane glycoproteins.
Antigenic properties
The alphaviruses were originally described as Group A arboviruses based upon their antigenic cross-relationships. Using specific serological testing, antigenic complexes were developed where all members of a particular complex were closely related to each other. Eight (nine including the fish alphaviruses) such complexes are described whose members, for the most part, are also genetically clustered. The members of the two genera of the Togaviridae are antigenically quite distinct from each other with no detectable antigenic homology.
Biological properties
Alphaviruses are, for the most part, arthropod-borne viruses which replicate in both their invertebrate vectors and their vertebrate hosts. (See Figure 3.) The salmonid alphaviruses and southern elephant seal virus, which cause disease in several species of fish and marine mammals, are exceptions because they seemingly lack an invertebrate vector. Similarly, RUBV appears to only infect mammals via the respiratory route but tends to develop persistent infections in contrast to the alphaviruses which are cytopathic to vertebrate cells. Both alphaviruses and rubiviruses have a global distribution and are capable of causing significant human illness. RUBV causes a mild febrile illness in children and adults but can cause significant teratology if a woman is infected during her first trimester. Alphaviruses cause disease ranging from mild febrile illness to prolonged arthritis to encephalitis.
Genus Alphavirus
Type species Sindbis virus
Distinguishing features
Alphaviruses are transmitted between vertebrate hosts via the bite of a persistently infected arthropod vector, most frequently a mosquito. Each distinct virus has a preferred invertebrate vector and infection of the vector is typically lifelong and non-cytopathic. Vertebrate hosts can also vary significantly with distinct viruses infecting mammals, birds, fish, reptiles and amphibians. Geographically, alphaviruses span the globe yet many of the viruses are found in highly limited geographic areas with specific ecological conditions suited to their survival. Severe disease (encephalitis) is associated primarily with New World geography while febrile illness and arthralgia are more globally distributed. While the overriding common characteristics are genome organization and arthropod-borne status, the alphaviruses are a widely diverse group of viruses in virtually all other characteristics.
Species demarcation criteria in the genus
Distinct species within the same antigenic complex generally show at least 21% nucleotide and 8% amino acid sequence divergence. Viruses in different serocomplexes typically diverge by over 38% at the nucleotide level and 40% at the amino acid level. Members of different species usually have distinct ecological and transmission patterns.
List of species in the genus Alphavirus
| Aura virus |
|
|
| Aura virus BeAR 10315 | [AF126284] | (AURAV-BeAR10315) |
| Barmah Forest virus |
|
|
| Barmah Forest virus BH2193 | [U73745] | (BFV-BH2193) |
| Bebaru virus |
|
|
| Bebaru virus MM2354 | [U94595*, AF339480*] | (BEBV-MM2354) |
| Cabassou virus |
|
|
| Cabassou virus CaAr 508 | [AF075259] | (CABV-CaAr508) |
| Chikungunya virus |
|
|
| Chikungunya virus 37997 | [AY726732] | (CHIKV-37997) |
| Eastern equine encephalitis virus |
|
|
| Eastern equine encephalitis virus BeAr436087 | [EF151503] | (EEEV-BeAr436087) |
| Everglades virus |
|
|
| Everglades virus Fe3-7c | [AF075251] | (EVEV-Fe37c) |
| Fort Morgan virus |
|
|
| Fort Morgan virus CM4-146 | [GQ281603] | (FMV-CM4-146) |
| Buggy Creek virus 81V8122 | [AF339474*, U94607*, U60403*] |
|
| Getah virus |
|
|
| Getah virus M1 | [EU015061] | (GETV-M1) |
| Highlands J virus |
|
|
| Highlands J virus B-230 | [GQ227789] | (HJV-B230) |
| Mayaro virus |
|
|
| Mayaro virus Brazil | [AF237947] | (MAYV - Brazil) |
| Middelburg virus |
|
|
| Middelburg virus MIDV857 | [EF536323] | (MIDV-857) |
| Mosso das Pedras virus |
|
|
| Mosso das Pedras virus 78V3531 | [AF075257] | (MDPV-78V3531) |
| Mucambo virus |
|
|
| Mucambo virus BeAn 8 | [AF075253] | (MUCV-BeAn8) |
| Ndumu virus |
|
|
| Ndumu virus SaAr 2204 | [U94600*, AF339487*] | (NDUV-SaAr2204) |
| O’nyong-nyong virus |
|
|
| O’nyong-nyong virus SG650 | [AF079456] | (ONNV-SG650) |
| Pixuna virus |
|
|
| Pixuna virus BeAr35645 | [AF075256] | (PIXV-BeAr35645) |
| Rio Negro virus |
|
|
| Rio Negro virus AG80-663 | [AF075258] | (RNV-AG80-663) |
| Ross River virus |
|
|
| Ross River virus NB5092 | [M20162] | (RRV-NB5092) |
| Sagiyama virus | [AB032553] |
|
| Salmon pancreas disease virus |
|
|
| Salmon pancreas disease virus F93125 | [AJ316244] | (SPDV-F93125) |
| Sleeping disease virus | [AJ316246] |
|
| Semliki Forest virus |
|
|
| Semliki Forest virus 42S | [X04129] | (SFV-42S) |
| Sindbis virus |
|
|
| Babanki virus DakAry 251 | [U94604*, AF339477*] |
|
| Kyzylagach virus LEIV 65A | [U94605*, AF339478*] |
|
| Ockelbo virus Edsbyn | [M69205] |
|
| Sindbis virus hssp | [J02363] | (SINV-hssp) |
| Southern elephant seal virus |
|
|
| Southern elephant seal virus Macquarie Island | [AF315122*] | (SESV-McQI) |
| Tonate virus |
|
|
| Tonate virus CaAn 410d | [AF075254] | (TONV-CaAn410d) |
| Trocara virus |
|
|
| Trocara virus BeAr422431 | [AF252265*] | (TROV-BeAr422431) |
| Una virus |
|
|
| Una virus BeAr 13136 | [U94603*, AF339481*] | (UNAV-BeAr13136) |
| Venezuelan equine encephalitis virus |
|
|
| Venezuelan equine encephalitis virus Trinidad donkey | [L01442] | (VEEV-TRD) |
| Western equine encephalitis virus |
|
|
| Western equine encephalitis virus McMillan | [GQ287640] | (WEEV-McMillan) |
| Whataroa virus |
|
|
| Whataroa virus M78 | [U94606*, AF339479*] | (WHAV-M78) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Alphavirus but have not been approved as species
None reported.
Genus Rubivirus
Type species Rubella virus
Distinguishing features
In contrast to the alphaviruses which are arthropod-borne, RUBV is transmitted by aerosol. The illness, known as rubella or German measles, generally consists of fever and rash; complications are rare. However, if a pregnant woman is infected with RUBV during the first trimester there is a 20% chance of the fetus developing congenital rubella syndrome (CRS). Birth defects due to CRS include deafness, cataracts, heart defects, mental retardation, and liver and spleen damage.
CRS infants shed virus for up to six months. Such persistence contrasts with alphavirus infections which are cleared within days or weeks. RUBV is endemic worldwide and is vaccine preventable.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Rubivirus
| Rubella virus |
|
|
| Rubella virus RA 27/3 | [L78917] | (RUBV-RA27/3) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Rubivirus but have not been approved as species
None reported.
List of unassigned species in the family Togaviridae
None reported.
Phylogenetic relationships within the family
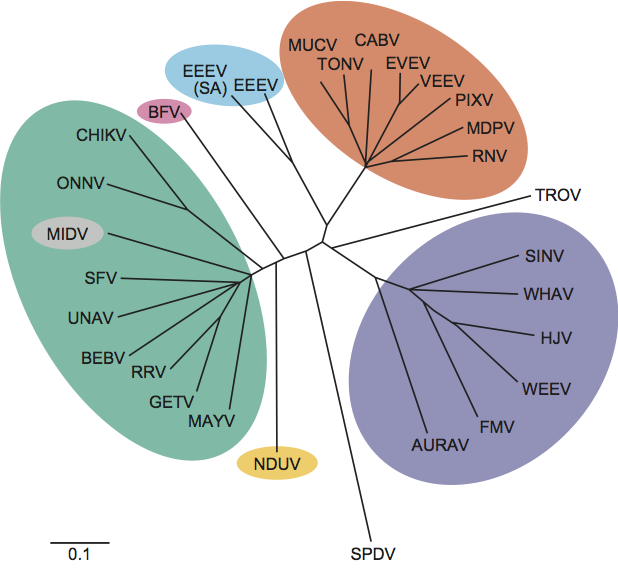
The alphaviruses have been extensively sequenced and phylogenetic comparisons made using whole genomes, E1, nsP1 and nsP4 genes. Distinct species within the same antigenic complex generally show at least 21% nucleotide and 8% amino acid sequence divergence. Viruses in different serocomplexes typically diverge by over 38% at the nucleotide level and 40% at the amino acid level. RUBV is quite distinct genetically from the alphaviruses demonstrating amino acid homology only within the non-structural proteins. (See Figure 4.)
Similarity with other taxa
Flaviviruses were previously included in the family Togaviridae suggesting some biological and virological similarities to the current members of this family. An atomic resolution crystal structure of an alphavirus E1 protein shows a folding pattern related to the E protein of flaviviruses, suggesting homology of at least some genes between these families. However, distinct genetic organization and replication led to the separation of these groups. There has also been the suggestion that similarities in replication proteins among members of RNA viruses from several plant families could imply an alphavirus-like superfamily exists.
Derivation of names
Alpha: from Greek letter α.
Rubi: from Latin rubeus, “reddish”.
Toga: from Latin toga, “cloak”.
Further reading
Fringuelli et al., 2008 E. Fringuelli, H.M. Rowley, J.C. Wilson, R. Hunter, H. Rodger, D.A. Graham, Phylogenetic analyses and molecular epidemiology of European salmonid alphaviruses (SAV) based on partial E2 and nsP3 gene nucleotide sequences. J. Fish Dis. 31 (2008) 811–823.
Griffin, 2007 D.E. Griffin, D.M. Knipe, P.M. Howley, D.E. Griffin, R.A. Lamb, M.A. Martin, B. Roizman, S.E. Strauss, AlphavirusesFields Virology. In: D.M. Knipe, P.M. Howley, D.E. Griffin, R.A. Lamb, M.A. Martin, B. Roizman, S.E. Strauss, Fields Virology. Lippincott, Williams and Wilkins, Philadelphia, PA20071023–1067.
Hobman and Chantler, 2007 T. Hobman, J. Chantler, D.M. Knipe, P.M. Howley, D.E. Griffin, R.A. Lamb, M.A. Martin, B. Roizman, S.E. Strauss, Rubella virusFields Virology. In: D.M. Knipe, P.M. Howley, D.E. Griffin, R.A. Lamb, M.A. Martin, B. Roizman, S.E. Strauss, Fields Virology. Lippincott, Williams and Wilkins, Philadelphia, PA20071069–1100.
Karabatsos, 1985 N. Karabatsos, International Catalog of Arboviruses Including Certain Other Viruses of Vertebrates. American Society of Tropical Medicine and Hygiene, San Antonio, TX1985.
Kuhn, 2007 R.J. Kuhn, D.M. Knipe, P.M. Howley, D.E. Griffin, R.A. Lamb, M.A. Martin, B. Roizman, S.E. Strauss, Togaviridae: The viruses and their replicationFields Virology. In: D.M. Knipe, P.M. Howley, D.E. Griffin, R.A. Lamb, M.A. Martin, B. Roizman, S.E. Strauss, Fields Virology. Lippincott, Williams and Wilkins, Philadelphia, PA20071001–1022.
Mukhopadhyay et al., 2006 S. Mukhopadhyay, W. Zhang, S. Gabler, P.R. Chipman, E.G. Strauss, J.H. Strauss, T.S. Baker, R.J. Kuhn, M.G. Rossmann, Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure. 14 (2006) 63–73.
Pfeffer et al., 1998 M. Pfeffer, R.M. Kinney, O.R. Kaaden, The alphavirus 3′-nontranslated region: size heterogeneity and arrangement of repeated sequence elements. Virology. 240 (1998) 100–108.
Powers et al., 2001 A.M. Powers, A.C. Brault, Y. Shirako, E.G. Strauss, W. Kang, J.H. Strauss, S.C. Weaver, Evolutionary relationships and systematics of the alphaviruses. J. Virol. 75 (2001) 10118–10131.
Strauss and Strauss, 1994 J.H. Strauss, E.G. Strauss, The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58 (1994) 491–562.
Contributed by
Powers, A., Huang, H., Roehrig, J., Strauss, E. and Weaver, S.
Figures
Figure 1 Thin section electron micrographs of Chikungunya virus in Vero E6 cells. Infected cells showed an abundance of viral particles that tended to associate with the plasma membrane.
(Courtesy of Cynthia Goldsmith, MS and James A. Comer, PhD, CDC.)
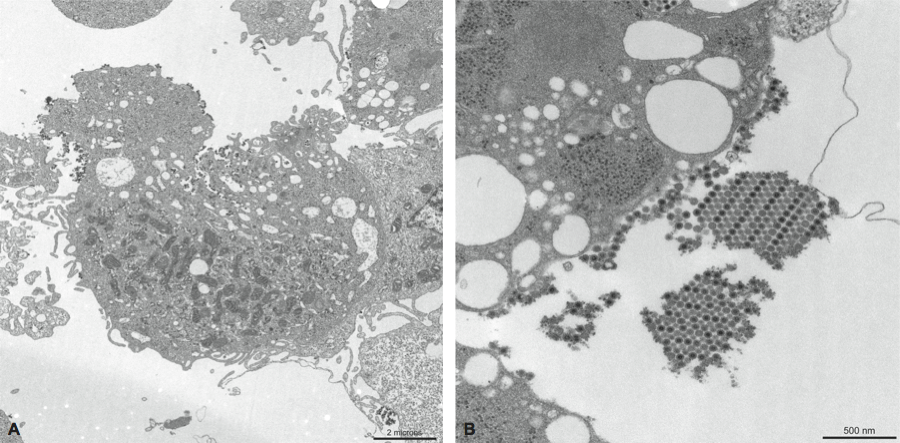
Figure 2 Togavirus genomic coding strategies. Shown are comparative schematic representations of the alphavirus and rubivirus genomic RNAs with UTRs represented as solid black lines and ORFs as open boxes (NS-ORF=non-structural protein ORF; S-ORF=structural protein ORF). Within each ORF, the coding sequences for the proteins processed from the translation product of the ORF are delineated. The asterisk between nsP3 and nsP4 in the alphavirus NS-ORF indicates the stop codon present in some alphaviruses that must be translationally read through to produce a precursor containing nsP4. Additionally, within the NS-ORFs, the locations of motifs associated with the following activities are indicated: (Mtr) methyl transferase, (Pro) protease, (Hel) helicase, (X) unknown function, and (Rep) replicase. The sequences encompassed by the sgRNA are also shown.
(Courtesy of T. K. Frey.)
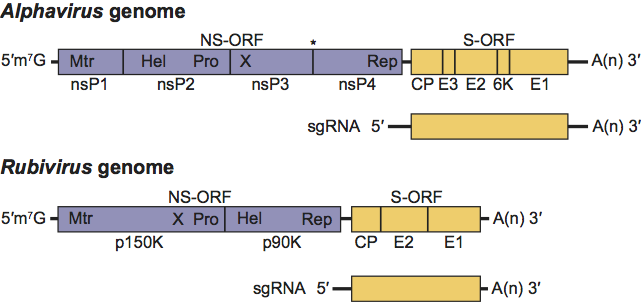
Figure 3 Model for the processing of the alphavirus nonstructural polyprotein during replication. When low levels of P123 are present, cis-cleavage of P1234 generates the minus-strand RNA replicase of the virus. This results in primarily negative sense RNA being generated from the incoming genomic RNA of the virus (upper panel). As the level of the trans-acting protease P123 rises in the infected cell, cleavage in trans generates other RNA replicase complexes. This results in a shift by the virus from the production of primarily negative sense RNA to primarily positive sense RNA. Eventually, replicase complexes capable of producing negative sense RNA will no longer be present in the infected cell resulting in the complete cessation of negative sense RNA synthesis (lower panel). The presence of a leaky opal termination codon (depicted as a black diamond) in the virus genome is believed to lead to a more rapid buildup of P123 in the infected cell, and thus a more rapid conversion to the production of positive sense RNA by the virus.
(Courtesy of Dr Kevin Myles; modified from Powers, A.M. (2008). Togaviruses: Alphaviruses. In: Encyclopedia of Virology, 3rd edn (B.W.J. Mahy and M.H.V. Van Regenmortel, Eds.), Oxford, Elsevier, pp. 96-100.)
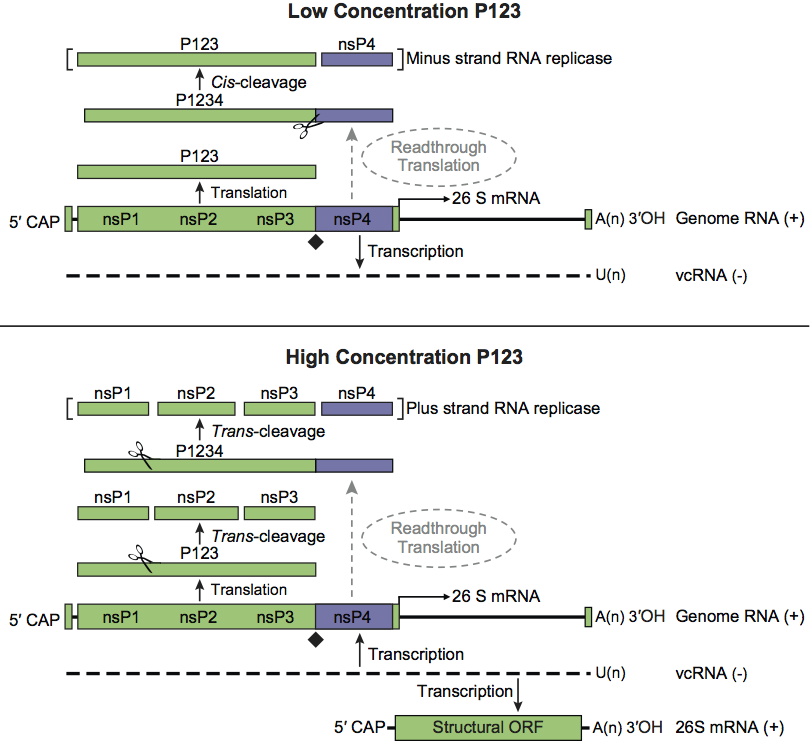
Figure 4 Unrooted phylogenetic tree of representative isolates of all alphavirus species generated from the E1 nucleotide sequences using the F84 algorithm of the neighbor-joining program (SESV and RUBV are not included because no homologous sequence for this region is available). Virus abbreviations are from the list of species in the genus Alphavirus. Antigenic complexes are indicated by colored circles.
(From Powers, A.M. (2008). Togaviruses: Alphaviruses. In: Encyclopedia of Virology, 3rd edn (B.W.J. Mahy and M.H.V. Van Regenmortel, Eds.), Oxford, Elsevier, pp. 96-100.)