Family: Tetraviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
NOTE: In 2011 the family Tetraviridae was divided into three unassigned families: Alphatetraviridae, Carmotetraviridae and Permutotetraviridae. The chapter below, reflects the taxonomy at the time of the 2009 Taxonomy release.
Genus Betatetravirus
Type species Nudaurelia capensis beta virus
Virion properties
Morphology
Virions are non-enveloped, roughly spherical, about 40 nm in diameter and exhibit T=4 icosahedral shell quasi-symmetry. Distinct capsomers have been resolved by cryo-electron microscopy and image reconstruction (Figure 1). The genome consists of ssRNA. Viruses in the genus Betatetravirus have monopartite genomes.
Physicochemical and physical properties
Virion Mr is about 18×106. Virion S20,w is 194–217S. Virion buoyant density in CsCl is usually 1.28–1.30 g cm−3 but occasionally as high as 1.33 g cm−3 (varies with species). Virions are stable over a broad range of pH and their infectivity can resist desiccation and protease treatment.
Nucleic acid
Virions of Nudaurelia capensis beta viruses (NβV), which form the type species Nudaurelia capensis beta virus, contain a single, positive sense, ssRNA segment of approximately 6.6 kb (Mr 1.8×106) which represents about 10% of the particle mass. This genomic RNA (gRNA) is not polyadenylated at its 3′ end, nor blocked like nodaviral RNAs, but terminates instead with a distinctive tRNA-like structure. A pseudoknot is found instead at the 3′ ends of the gRNA of Euprosterna elaeasa virus (EeV) and Thosea asigna virus (TaV) (prototypes of species of the same name). These structures are absent at the 3′ end of the gRNAs of Providence virus (PrV; Providence virus prototype). A subgenomic message for the capsid proteins (CPs), which corresponds to the 3′ end of the genomic RNA, can also be encapsidated in some species.
Proteins
Capsids of NβV consist of 240 protein subunits (protomers) arranged on a T=4 surface lattice. Each protomer consists of the two cleavage products (large, L or β, 58.4 kDa and small, S or γ, 8 kDa), of a single CP precursor (α 66.4 kDa). Minor amounts of the un-cleaved precursor may be found in virions. Capsids of TaV, EeV, and PrV have similar protein compositions.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Betatetraviruses replicate in the cytoplasm. Three distinct types of genomic organization have so far been found for betatetraviruses. The 6,625 nt gRNA of NβV, contains two ORFs that overlap for 1,517 nt: the first ORF at the 5′ end contains 1,821 codons and encodes the putative multidomain RNA replicase (204 kDa), which almost entirely overlaps the second ORF of 612 codons at the 3′ end that encodes the capsid protein precursor (66.4 kDa) (Figure 2). The replicase includes three highly conserved enzymatic domains, N7-methyltransferase (NMT), superfamily 1 RNA helicase (HEL1) and RNA-dependent RNA polymerase typical of the alpha-like supergroup (acRdRp) that are yet to be characterized experimentally.
The second type of gRNA is represented by the closely related TaV and EeV (5,714 and 5,698 nt, respectively). At the 5′ end is the longer ORF (1,257 codons) encoding the putative replicase of approximately 140 kDa. The viral replicase carries an RdRp domain with a non-canonical, permuted, CAB motif arrangement (pRdRp). At the N-terminus this domain is flanked by a short conserved signal implicated in priming replication and covalent binding to the 5′ end of RNAs (virus protein of genome, VPg). Two conserved domains flanking VPg-pRdRp have no resemblance to nmT or HEL1 domains. There is a much shorter overlap (536 and 529 nts for EeV and TaV, respectively) between the replicase and CP ORFs compared to that of NβV. The TaV and EeV CP ORFs are longer than that of NβV, yielding a putative precursor of 757 aa in length that undergoes NPGP-mediated (2A-like) processing to produce an amino terminal portion of 17 kDa (Figure 2) in addition to the capsid protein precursor of 65 kDa.
The third type of genomic organization is represented by PrV gRNA that has an intermediate size of 6,155 nt (Figure 2). At the 5′ end is an ORF of 3,659 nt comprising 1,233 amino acids encoding a protein of 130 kDa, of unknown function (ORF1) that includes an NPGP (2A-like) signal at the N-terminus. ORF1 (p130) almost completely overlaps with ORF2, encoding the viral replicase (2,934 nt, 978 amino acids and a protein of 104 kDa). ORF2 contains a readthrough stop codon that attenuates the synthesis of 104 kDa protein to produce a 40 kDa peptide from its N-terminus. The canonical RdRp domain, which is typical of the carmo-like supergroup, (ccRdRp) resides in the C-terminal half of the replicase, downstream of the readthrough stop and there is no evidence of nmT or HEL1 domains. The CP ORF is located immediately downstream of the replicase ORF, comprising 2,262 nt and 754 amino acids that undergoes NPGP-mediated (2A-like) processing at the second of two sites in the amino terminus of the protein, producing a peptide of 15 kDa and the 68 kDa capsid protein precursor.
During RNA replication of NβV, TaV and PrV, a subgenomic RNA (sgRNA), which represents the 3′ 2,500 nt of the genome is synthesized, and this serves as the mRNA for the CP precursor (Figure 2). There is a suggestion that the PrV sgRNA may replicate independently, and if confirmed, this, rather than the packaging of sgRNAs might blur the distinction between the monopartite genome organization of the betatetraviruses and the bipartite genome organization of the omegatetraviruses.
Antigenic properties
Most of the members of the group are serologically interrelated but distinguishable. The majority of the isolates were identified on the basis of their serological reaction with antiserum raised against NβV.
Biological properties
Host range
Nature: All virus species were isolated from Lepidoptera species (moths and butterflies), principally from Saturniid, Limacodid and Noctuid moths and no replication in other animals has been detected. In larvae, virus replication is restricted predominantly to the cells of the midgut.
Laboratory: With the exception of PrV, no infections by members of the Betatetravirus genus have been achieved in cultured cells, even when gRNA was transfected directly into cells.
Transmission
Oral transmission of NβV to Antherea eucalypti (the emperor gum moth) has been demonstrated experimentally. Oral transmission is implied by the midgut site of viral replication and by reports of some tetraviruses being used as sprayed insecticides in Malaysia (e.g. DtV; Darna trima virus). At high host densities, horizontal spread appears to be the major route of infection. Suggestive evidence exists for vertical transmission, which could be responsible for the observed persistence of tetraviruses within insect populations.
Cytopathic effects
The viruses replicate primarily in the cytoplasm of gut cells of several Lepidoptera species. Crystalline arrays of virus particles are often seen within cytoplasmic vesicles. Different isolates vary considerably in pathogenicity; symptoms can vary from inapparent to acutely lethal infections.
Species demarcation criteria in the genus
Viruses have been classified into the genus Betatetravirus based upon two properties: capsid of T=4 symmetry and monopartite genome. The following criteria have been applied to the demarcation of species within the genus:
- Biological properties (host range, vectors, mode of transmission). Since the natural host-ranges of individual recognized tetravirus species appear to be narrow, virus isolation from a new host can provide suggestive evidence of a new tetravirus species.
- Antigenic properties. Antisera raised against different isolates or strains of a single tetravirus species should exhibit high levels of cross-reactivity in Western blot and/or neutralization analyses. Lower levels of cross-reactivity in these assays using antisera against previously recognized tetraviruses can provide evidence of a new tetravirus species.
- Virion physical/physicochemical characteristics. In the absence of more definitive criteria, significant (>5%) differences in virion sedimentation coefficient or buoyant density from those of all previously recognized tetravirus species can provide evidence of a new virus species.
- Structural protein characteristics. The electrophoretic mobilities in SDS-PAGE of the CP precursor or its cleavage products should be compared with those of other tetravirus species.
- Genome molecular characteristics:
- RNA electrophoretic mobilities. In the absence of sequence information, the electrophoretic mobilities of the viral genomic RNAs should be compared with those of other tetraviruses.
- RNA hybridization properties. In the absence of differences in RNA electrophoretic mobilities, the molecular hybridization properties of the viral genomic RNAs should be compared with those of other tetraviruses.
- Genome sequence characteristics. The nt sequences of the genomic RNA(s) should be compared with those of other tetraviruses.
Application of these criteria: In practice, while criteria 1–5 above may be suggestive of a new species, definitive demarcation has been based on the nucleotide sequence of the viral CP gene. Application of the sixth criterion to viruses of this genus led to suggestions, based on profound differences in the replicase genes accounting for a considerable portion of the genome, that TaV, EeV and PrV should be reclassified outside the Tetraviridae. A taxonomic proposal to this effect is currently under consideration.
List of species in the genus Betatetravirus
| Antherea eucalypti virus |
|
|
| Antherea eucalypti virus |
| (AeV) |
| Darna trima virus |
|
|
| Darna trima virus |
| (DtV) |
| Dasychira pudibunda virus |
|
|
| Dasychira pudibunda virus |
| (DpV) |
| (Calliteara pudibunda virus) |
| (CpV) |
| Euprosterna elaeasa virus |
|
|
| Euprosterna elaeasa virus | [AF461742] | (EeV) |
| Nudaurelia capensis beta virus |
|
|
| Nudaurelia capensis β virus | [AF102884] | (NβV) |
| Philosamia cynthia x ricini virus |
|
|
| Philosamia cynthia x ricini virus |
| (PxV) |
| Providence virus |
|
|
| Providence virus | [GU991616] | (PrV) |
| Pseudoplusia includens virus |
|
|
| Pseudoplusia includens virus |
| (PiV) |
| Thosea asigna virus |
|
|
| Thosea asigna virus | [AF062037*, AF282930*] | (TaV) |
| (Setothosea asigna virus) |
| (SaV) |
| Trichoplusia ni virus |
|
|
| Trichoplusia ni virus |
| (TnV) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Betatetravirus but have not been approved as species
None reported.
Genus Omegatetravirus
Type species Nudaurelia capensis omega virus
Virion properties
Morphology
Virions are non-enveloped, roughly spherical, about 40 nm in diameter and exhibit T=4 icosahedral shell quasi-symmetry. Distinct capsomers have been resolved by cryo-electron microscopy and image reconstruction (Figure 3). The genome consists of ssRNA. Unlike viruses in the genus Betatetravirus, viruses in the genus Omegatetravirus have bipartite genomes.
Physicochemical and physical properties
Virion Mr is about 16×106. Virion S20,w is 194–217S. Virion buoyant density in CsCl is 1.28–1.30 g cm−3 (varies with species). Virions of Helicoverpa armigera stunt virus (HaSV) are stable between pH 3.0 and 11.0 and at temperatures up to 55 °C and are resistant to protease treatment.
Nucleic acid
Virions of viruses of the type species Nudaurelia capensis omega virus that have the same name and commonly abbreviated as NωV, contain two positive sense, ssRNA segments of approximately 5,300 nt (RNA1, Mr 1.75×106) and 2,450 nt (RNA2, Mr 0.8×106). These genomic RNAs are capped at their 5′ ends but their 3′ ends are not polyadenylated, nor blocked. Like NβV, omegatetraviral RNAs terminate with a distinctive tRNA-like structure. It is likely that omegatetraviruses encapsidate both genomic RNAs within a single particle, in which case the RNAs would represent about 10% of the particle mass.
Proteins
Capsids of the type strain, NωV, consist of 240 protein subunits (protomers) arranged on a T=4 surface lattice. Each protomer consists of the two cleavage products (L or β, 62 kDa and S or γ, 7.8 kDa) of a single CP precursor (α, 69.8 kDa). Overall, the CPs of NωV and NβV share less than 20% aa sequence identity, indicating that viruses of the two genera have substantially diverged with respect to their capsid sequences.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
As with the betatetraviruses, omegatetraviruses replicate in the cytoplasm. Studies in tissue culture cells show that the HaSV replicase is localized within the cytoplasm and associates with membranes derived from the endocytic pathway. The viral genome consists of two unique molecules of RNA, probably encapsidated in the same virus particle. The NωV genome has not yet been fully sequenced but the partial sequence available for RNA1 indicates that this virus has a similar genome organization to that of two other omegatetraviruses that have been fully characterized in this respect. These omegatetraviruses form separate species with identical or similar names (Table 2). RNA1 (HaSV: Mr 1.75×106; 5.3 kb), encodes the RNA replicase, 1,704 aa and 1,649 aa in HaSV and Dendrolimus punctatus tetravirus (DpTV), respectively, with a domain organization similar to that of the betatetravirus NβV (Figure 1). RNA1 also encodes three small ORFs of unknown function that overlap with the 3′ end of the replicase ORF. RNA2 (HaSV: Mr 0.8×106; 2.45 kb) encodes the CP precursor (70 kDa for NωV and DpTV and 71 kDa for HaSV). A 157-codon ORF precedes and partially overlaps the CP ORF; it encodes a non-structural protein (p17), which is encapsidated and involved in packaging of viral RNA. Separate dsRNAs that correspond to the two genome segments are observed in HaSV-infected cells.
Antigenic properties
NωV and DpTV are serologically related, but display no serological relationship with more distantly related HaSV.
Biological properties
Host range
Nature: All species were isolated from Lepidoptera species, principally from Saturniid, Limacodid and Noctuid moths.
Laboratory: No infections by members of the genus Omegatetravirus have yet been achieved in cultured cells, but infectious HaSV particles were produced by plant protoplasts transfected with plasmids carrying full-length cDNAs that corresponded to the viral genome segments.
Transmission
As with the betatetraviruses, oral transmission is implied by the midgut site of viral replication. At high host densities, horizontal spread appears to be the major route of infection, but evidence exists for vertical transmission which might be responsible for the observed persistence of tetraviruses within insect populations.
Cytopathic effects
The viruses replicate primarily in the cytoplasm of midgut cells of the larvae of several Lepidoptera species. Crystalline arrays of virus particles are often seen within cytoplasmic vesicles. There is a considerable range of pathogenicity with different isolates, and symptoms can vary from unapparent to acutely lethal infections.
Species demarcation criteria in the genus
Viruses have been classified into the genus Omegatetravirus based upon two properties: capsid of T=4 symmetry and bipartite genome. The species demarcation criteria of betatetraviruses also apply to omegatetraviruses. Also, because the genome of omegatetraviruses is segmented, re-assortment is possible and the two genome segments may have different evolutionary histories. However, no chimeric omegatetraviruses have yet been detected. In contrast, it was proposed that the ancestral bi-segmented omegatetravirus has evolved by recombination from two mono-segmented betatetraviruses of the NβV and PrV types, respectively. This proposal suggested that the NβV-like parent supplied genomic RNA encoding the replicase, while the other PrV-like parent supplied a subgenomic RNA encoding CP, which subsequently evolved into the second segment by mutation and autonomization.
List of species in the genus Omegatetravirus
| Dendronimus punctatus virus |
|
|
| Dendronimus punctatus tetravirus | RNA1 [AY594352] | (DpTV) |
|
| RNA2 [AY594353] |
|
| Helicoverpa armigera stunt virus |
|
|
| Helicoverpa armigera stunt virus | RNA1 [U18246] | (HaSV) |
|
| RNA2 [L37299] |
|
| Nudaurelia capensis omega virus |
|
|
| Nudaurelia capensis ω virus | RNA2 [S43937] | (NωV) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Omegatetravirus but have not been approved as species
None reported.
List of other related viruses which may be members of the family Tetraviridae but have not been approved as species
| Acherontia atropas virus | (AaV) |
| Agraulis vanillae virus | (AvV) |
| Callimorpha quadripuntata virus | (CqV) |
| Eucocytis meeki virus | (EmV) |
| Euploea corea virus | (EcV) |
| Hyalophora cecropia virus | (HcV) |
| Hypocritae jacobeae virus | (HjV) |
| Lymantria ninayi virus | (LnV) |
| Saturnia pavonia virus | (SpV) |
| Setora nitens virus | (SnV) |
| Nudaurelia capensis virus* | (NV) |
| Nudaurelia capensis ζ virus | (NζV) |
| Nudaurelia capensis ψ virus | (NψV) |
* NV resembles the tetraviruses in appearance but is serologically unrelated to any known species.
Phylogenetic relationships within the family
Two major clusters are evident in the tetravirus capsid phylogram, with one comprising NβV, TaV and EeV, and the other DpTV, HaSV, NωV and PrV (Figure 4). HaSV, NωV and DpTV appear to have evolved more recently, sharing an early common ancestor with PrV, while the closely related TaV and EeV share a most recent common ancestor with NβV. It is striking that in each of these clusters, both CP processing strategies, with and without involvement of the NPGP signal, are present.
In contrast to the CPs, the replicases of the tetraviruses for which genomic sequences have been determined fall into three distinct phylogenetic groups that do not reflect the taxonomic demarcation (Figure 5). The first of these groups includes the betatetravirus NβV and the omegatetraviruses HaSV, DpTV and probably also NωV (although no complete sequence has been published for the replicase of NωV, unpublished data show it to be very closely related to that of HaSV). Replicases of these viruses include three conserved domains, nmT, HEL1 and acRdRp, and cluster together with the similarly organized replicases of a dozen other ssRNA+ virus families with the mammalian Hepeviridae family being the closest (Figure 5; see also below). The second group includes the betatetraviruses TaV and EeV. The replicases of these viruses are also multi-domain proteins with VPg and pRdRp being two domains with provisionally assigned functions. The phylogenetic neighborhood of the TaV, EeV pRdRps includes diverse viruses that are discussed below but not those of the NβV/HaSV group (Figure 6). There is a third distinct RdRp lineage within the tetraviruses represented by PrV, whose RdRp clusters with ssRNA+ plant viruses distinct from those that form either of the above two groups (Figure 7).
Similarity with other taxa
The distinguishing feature of viruses currently classified within the family Tetraviridae is the unique T=4 quasi-symmetry of their capsid architecture. Thus it is not surprising that their CPs form a monophyletic group. The jelly-roll fold β subunits of CPs, which are used to build capsid, are most closely but still distantly related to those of the CPs of nodaviruses and dsRNA birnaviruses having T=3 and T=13 capsids, respectively. It has been speculated that the tetravirus capsid might have evolved from a nodavirus-like ancestor through a process that included insertion of an immunoglobulin-like protein domain coding sequence (either acquired or evolved through sequence duplication) within the CP gene. This hypothesis is supported by structural studies that show that the PrV and NωV capsids are structurally conserved with respect to their jelly-roll folds and Ig-like domains, but not with respect to the structure and function of their N and C-termini. These domains, which form the molecular switch determining the T=4 architecture of PrV capsid, are closely related to those of nodaviruses that possess a T=3 capsid architecture.
Comparative analysis of currently available non-structural protein sequences split tetraviruses into three distinct lineages, prototyped by NβV/HaSV, TaV/EeV and PrV, respectively, within three different virus supergroups. The replicases of NβV, DpTV and HaSV resemble those of the “alphavirus-like” supergroup, having the distinct nmT-HEL1-acRdRp domain organization and through phylogenetic clustering with viruses having these three domains. The replicases of TaV and EeV lack both nmT and HEL1 domains. Furthermore, their pRdRp domain has a unique C-A-B motif arrangement in the palm subdomain of the active site that differs from the canonical A-B-C arrangement found in the other tetraviruses, all “alphavirus-like” viruses and indeed almost all known template-dependent polynucleotide polymerases (viral and cellular) carrying the palm sub-domain. Interestingly, the same C-A-B permutation of the three motifs is also found in replicases of all dsRNA birnaviruses. This rearrangement is a result of migration of about 22 aa residues encompassing motif C between two internal positions, separated by about 110 aa, in a conserved region of about 400 aa. The permuted TaV, EeV and birnavirus enzymes form a minor, deeply separated cluster in the RdRp tree that also includes viruses of the “picornavirus-like supercluster” and the order Nidovirales. The pRdRp domains of these viruses are also flanked by the uniquely conserved VPg signal and another poorly characterized domain from the N-terminus. A similar arrangement is also found in DAV. Thus, replicases of TaV/EeV and birnaviruses (and possibly DAV) may represent their own virus supercluster.
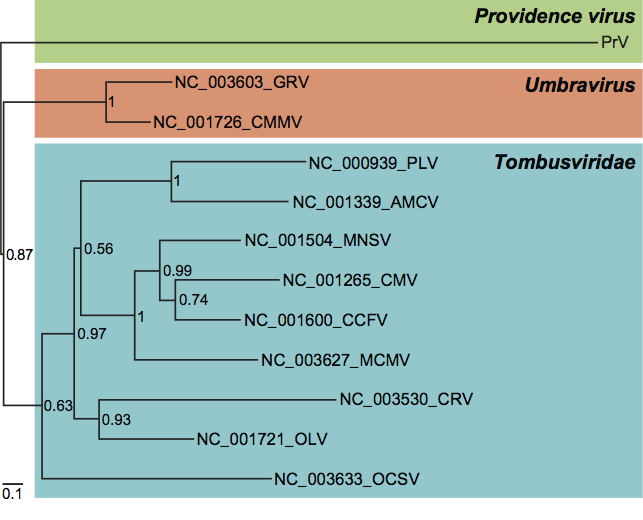
The RdRp of PrV also lacks the nmT and HEL1 domains, but unlike the TaV/EeV cluster, has a canonical A-B-C palm subdomain. The PrV replicase clusters with ssRNA+plant viruses, being most closely related to the umbra- and tombusviruses, which belong to the third virus supergroup, the carmo-like viruses of other families and therefore appears to belong to a third lineage.
These complex and incongruent relationships of CP and replicase proteins imply that viruses currently classified as tetraviruses on the basis of their CP may form a polyphyletic group based upon the properties of their replicase. According to a recently developed evolutionary model, TaV and EeV resemble the most recent common tetravirus ancestor while other tetraviruses descend from more recent ancestors that originated through recombination(s) with viruses of other families and/or intrafamily recombination. To acknowledge these gross differences, TaV/EeV and, probably PrV, are proposed to be placed in two new families separate from the prototypic tetraviruses. Future revision of tetravirus taxonomy is expected to accommodate these suggestions. A further question is whether additional families need to be defined to include viruses like DAV whose replicases are related to TaV/EeV but whose capsid is of the T=3 type.
Derivation of names
Nudaurelia capensis is the emperor pine moth.
Tetra: from Greek tettares, meaning “four”, as T=4.
Further reading
Brooks et al., 2002 E.M. Brooks, K.H.J. Gordon, S.J. Dorrian, E.R. Hines, T.N. Hanzlik, Infection of its lepidopteran host by the Helicoverpa armigera stunt virus (Tetraviridae). J. Invertebr. Pathol. 80 (2002) 97–111.
Dorrington and Short, 2010 R.A. Dorrington, J.R. Short, K. Johnson, S. Asgari, The TetravirusesThe Insect Viruses. In: K. Johnson, S. Asgari, The Insect Viruses. Horizon Scientific Press, UK2010.
Gorbalenya et al., 2002 A.E. Gorbalenya, F.M. Pringle, J.-L. Zeddam, B.T. Luke, C.E. Cameron, J. Kalmakoff, T. Hanzlik, K. Gordon, V.K. Ward, The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 324 (2002) 47–62.
Luke et al., 2008 G.A. Luke, P. de Felipe, A. Lukashev, S.E. Kallioinen, E.A. Bruno, M.D. Ryan, Occurrence, function and evolutionary origins of ‘2A-like’ sequences in virus genomes. J. Gen. Virol. 89 (2008) 1036–1042.
Pringle et al., 2003 F.M. Pringle, K.N. Johnson, C.L. Goodman, A.H. McIntosh, L.A. Ball, Providence virus: a new member of the Tetraviridae that infects cultured insect cells. Virology. 306 (2003) 359–370.
Short et al., 2010 J.R. Short, C. Knox, R.A. Dorrington, Subcellular localisation and live cell imaging of the Helicoverpa armigera stunt virus replicase in mammalian and Spodpoptera frugiperda cells. J. Gen. Virol. 91 (2010) 1514–1523.
Speir and Johnson, 2008 J.A. Speir, J.E. Johnson, B.W.J. Mahy, M.H.V. van Regenmortel, TetravirusesEncyclopedia of Virology. In: B.W.J. Mahy, M.H.V. van Regenmortel, Encyclopedia of Virology. Elsevier, Oxford200827–37.
Speir et al., 2010 J.A. Speir, D.J. Taylor, P. Natarajana, F.M. Pringle, L.A. Ball, J.E. Johnson, Evolution in action: N and C termini of subunits in related T=4 viruses exchange roles as molecular switches. Structure. 18 (2010) 700–709.
Walter et al., 2008 C.T. Walter, M. Tomasicchio, V. Hodgson, D.A. Hendry, M.P. Hill, R.A. Dorrington, Characterisation of a succession of small insect viruses in a wild South African population of Nudaurelia cytherea capensis. S. Afr. J. Sci. 104 (2008) 147–152.
Zeddam et al., 2010 J.-L. Zeddam, K.H.J. Gordon, C. Lauber, C.A. Felipe Alves, B.T. Luke, T.N. Hanzlik, V.K. Ward, A.E. Gorbalenya, Euprosterna elaeasa virus genome sequence and evolution of the Tetraviridae family: Emergence of bipartite genomes and conservation of the VPg signal with the dsRNA Birnaviridae family. Virology. 397 (2010) 145–154.
Contributed by
Dorrington, R.A., Gorbalenya, A.E., Gordon, K.H.J, Lauber, C. and Ward, V.K.
Figures
Figure 1 Betatetravirus capsid structure. (Left) Schematic representation of a T=4 icosahedral lattice. (Center and Right) Cryo-electron microscopy image reconstruction of a particle of Nudaurelia capensis virus (NV) on the symmetry axis 3 and 5; the bar represents 20 nm.
(Courtesy of H.R. Cheng, N. Olson and T. Baker.)
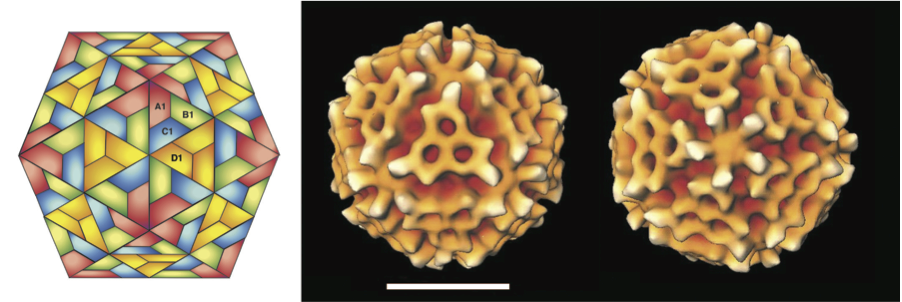
Figure 2 Tetravirus genome comparisons. The genome organization, including genome segments, ORFs and selected domains, is depicted for four tetraviruses, EeV, NV, HaSV and PrV, using virus-specific scales. EeV, NV and PrV have monopartite genomes that (are predicted to) yield sgRNAs, while HaSV has a bipartite genome (RNA1 and RNA2). The selected proteins and domains are labelled, pattern-coded and coloured to indicate homology. nmT: N7-methyltransferase; HEL1, superfamily 1 helicase; acRdRp: canonical RNA dependent RNA polymerase typical of alpha-like supergroup; ccRdRp: canonical RNA dependent RNA polymerase typical of carmo-like supergroup; pRdRp: non-canonical permuted RNA-dependent RNA polymerase most related to picorna-like supergroup; NPGP: 2A-like processing site. The sequence of the major (L or ) and minor (S or ) capsid proteins are indicated as L and S, respectively.
(Modified from Figure 1 of Zeddam et al. (2010). Virology, 397, 145-154.)
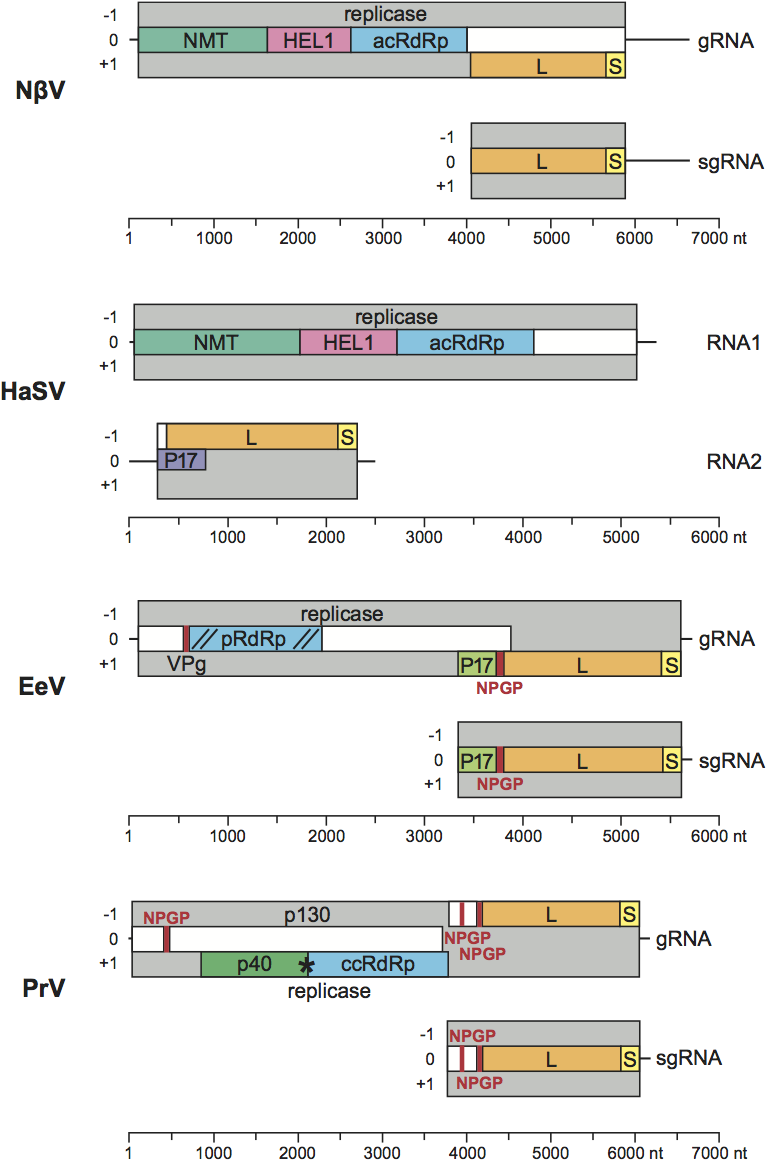
Figure 3 Comparison of tetravirus capsid morphologies. Cryo-electron microscopy reconstructions of capsids of the omegatetraviruses, Nudaurelia capensis virus (NV) and Helicoverpa armigera stunt virus (HaSV) and the betatetraviruses, NV and PrV. The structures, viewed down their two-fold axes, were determined at resolutions of between 25 and 30 .
(Image taken from Figure 2 of Speir and Johnson (2008); with permission.)
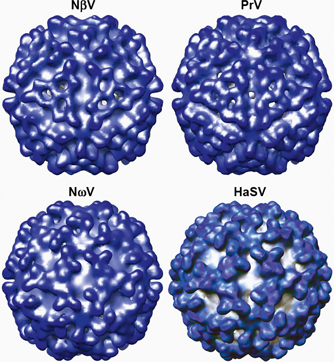
Figure 4 Phylogeny of tetravirus capsid protein. The tree for seven tetraviruses is based on an amino acid alignment of the jelly-roll domain of L protein (661 positions) and was rooted using the jelly-roll domain of S proteins of five birnaviruses as an outgroup. Numbers at branch points provide Bayesian posterior probability support values and the evolutionary scale is indicated by the bar of 0.5 amino acid substitutions per site on average.
(Modified from Figure 5b (left part) of Zeddam et al. (2010). Virology, 397, 145154.)
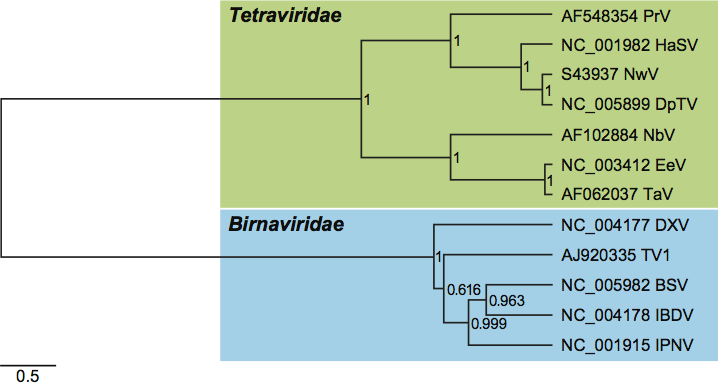
Figure 5 Phylogeny of prototypic tetravirus replicases. The tree for prototypic tetraviruses and DpTV is based on an amino acid alignment of the HEL1 and acRdRp domains (832 positions) and was rooted using avian and human Hepatitis E viruses as an outgroup. Numbers at branch points provide Bayesian posterior probability support values and the evolutionary scale is indicated by the bar of 0.5 amino acid substitutions per site on average.
(From Figure 5b (right part) of Zeddam et al. (2010). Virology, 397, 145154.)
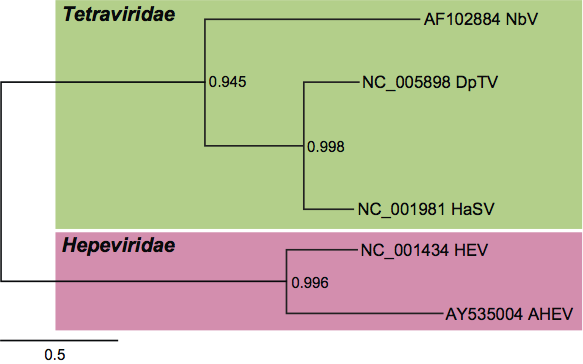
Figure 6 Unrooted phenogram showing the relationships of the RdRps of the tetraviruses TaV and EeV to other virus families and viruses in the picornavirus-like supercluster. The pRdRps of TaV, EeV and the birnaviruses were converted into the canonical form by relocating the motif C sequence (1820 aa) downstream of the motif B, as in canonical polymerase motifs. These sequences were aligned with those of polymerases from representative viruses in the Picornaviridae, Dicistroviridae, Secoviridae, Iflaviridae, Caliciviridae, Potyviridae, Coronavirinae, Torovirinae, Roniviridae, Arteriviridae and unclassified insect viruses. Using an extended, gap-free version of the alignment containing 332 informative characters, an unrooted neighbor-joining tree was inferred by the ClustalX1.81 program. All bifurcations with support in>700 out of 1000 bootstraps are indicated. Different groups of viruses are highlighted. Virus families and groups, viruses included in the analysis, abbreviations ( ) and the NCBI protein (unless otherwise specified) IDs [ ] are as follows: Picornaviridae: human poliovirus type 3 Leon strain (PV-3L) [130503] and human parechovirus 1 (HpeV-1) [6174922]; Iflaviridae: infectious flacherie virus (InFV) [3025415]; unclassified insect virus Acyrthosiphon pisum virus (APV) [7520835]; Dicistroviridae: Drosophila C virus (DCV) [2388673]; Secoviridae: rice tungro spherical virus (RTSV) [9627951], parsnip yellow fleck virus (PYFV) [464431], cowpea severe mosaic virus (CPSMV) [549316] and tobacco ringspot virus (TobRV) [1255221]; Caliciviridae: feline calicivirus F9 (FCV-F9) [130538] and Lordsdale virus (LORDV) [1709710]; Potyviridae: tobacco vein mottling virus (TVMV) [8247947] and barley mild mosaic virus (BaMMV) [1905770]; Coronavirinae: human coronavirus 229E (HCoV) [12175747]; Torovirinae: Berne torovirus (BEV) [94017]; Arteriviridae: equine arteritis virus (EAV) [14583262]; Roniviridae: gill-associated virus (GAV) [9082018]; Tetraviridae: Thosea asigna virus (TaV) [AF82930; nt sequence] and Euprosterna elaeasa virus (EeV) [AF461742; nt sequence]; Birnaviridae: infectious pancreatic necrosis virus (IPNV) [133634] and infectious bursal disease virus (IBDV) [1296811]. Coronaviridae, Arteriviridae and Roniviridae belong to the order Nidovirales.
(Modified from Gorbalenya et al., 2002.)
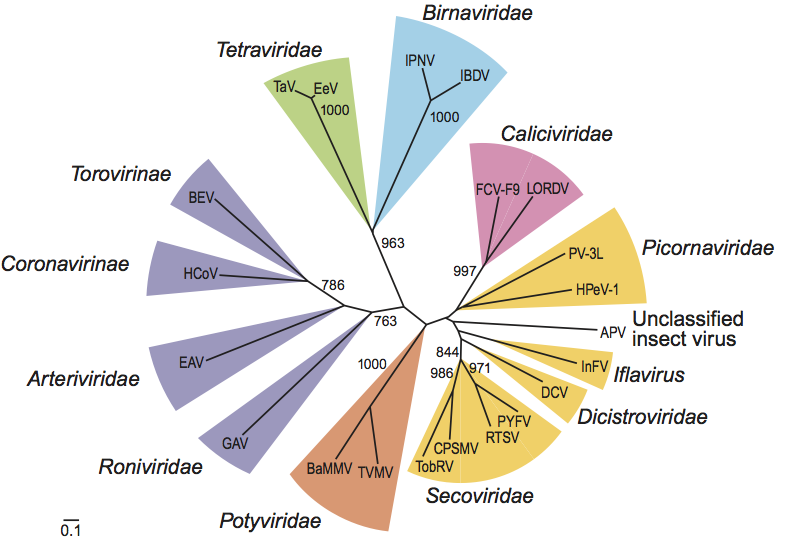
Figure 7 Phylogeny of Providence virus (PrV) and related tombus- and umbravirus RdRps. The tree is based on an amino acid alignment of the C-terminal half of the replicase starting after the read-through stop codon (578 positions) encoding putative RdRp and was midpoint pseudo-rooted. Poorly conserved alignment termini were discarded from the analysis. Tombus- and umbraviruses encode RdRps that are the closest to the PrV RdRp as determined in a protein Blast analysis. The RdRp tree was calculated and depicted following procedures and a style adopted by Zeddam et al. (2010) (see Figures 4 and 5). The relaxed lognormal molecular clock model allowing different branches of the tree to evolve at different rates was used. Numbers at branch points provide Bayesian posterior probability support values and the evolutionary scale is indicated by the bar of 0.1 amino acid substitutions per site on average. RefSeq accession numbers are indicated next to the virus names. This tree is by Lauber and Gorbalenya (unpublished) using the PrV sequence (GenBank accession no: AF548354).