Genus: Sobemovirus
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Type species: Southern bean mosaic virus
Distinguishing features
Sobemoviruses are similar in particle morphology, capsid stabilization, sedimentation coefficients, sizes of protein subunits and genomic RNA. All the sequenced sobemoviruses have a polycistronic positive sense single stranded RNA genome that consists of four ORFs. ORFs 1, 2a and 2b are all translated from the genomic RNA. Translation of ORF2a occurs via a leaky scanning mechanism. ORF2b is expressed as a fusion protein with ORF2a through a −1 ribosomal frameshift mechanism. The genome 3′-proximal ORF3 is translated from the subgenomic RNA.
Virion properties
Morphology
Virions are about 25–30 nm in diameter. They have a single tightly packed capsid layer with 180 subunits of about 26–31 kDa assembled on a T=3 icosahedral lattice. Sobemoviruses are stabilized by divalent cations, pH-dependent protein–protein interactions and salt bridges between protein and RNA. Upon alkali treatment in the presence of divalent chelators, the capsid swells and becomes sensitive to enzymes and denaturants. (See Figure 1.)
Physicochemical and physical properties
The virion Mr is about 6.6×106; S20,w is about 115S; density is about 1.36 g cm−3 in CsCl (but virus forms two or more bands in Cs2SO4); particles swell reversibly in EDTA and higher pH with concomitant changes in capsid conformation and partial loss of stability.
Nucleic acid
Particles contain a single molecule of positive sense ssRNA, about 4.0–4.5 kb in size. A subgenomic RNA molecule, co-terminal with the 3′ end of the genomic RNA, with a Mr of 0.3–0.4×106, is synthesized in the virus-infected cell. Both genomic and subgenomic RNAs are thought to have a viral genome-linked protein VPg covalently bound to their 5′ end. The 3′ terminus is non-polyadenylated and does not contain a tRNA-like structure. Several sobemoviruses encapsidate a circular viroid-like satellite RNA (220–390 nt).
Proteins
The major structural protein of sobemoviruses is the coat protein. In addition, from the central part of the polyprotein a viral genome-linked protein VPg is processed. It is predicted that sobemovirus VPgs are natively unfolded proteins.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
It has been presumed earlier that the genus includes viruses with two different genome organizations. Recently it has been demonstrated that these differences were due to sequencing errors and in fact the genus has the common genome organization depicted in Figure 2.
The protein P1, encoded by the 5′ terminal ORF of the genomic RNA, is the suppressor of gene silencing. It is essential for the systemic spread of the virus. The translation initiation codon of P1 has a poor context, enabling ribosomes to bypass it often and, by the utilization of the leaky scanning mechanism, initiate from the genomic RNA the translation of the next ORF. This ORF encodes the viral polyprotein. The first part of this ORF, ORF2a, encodes the virus serine protease, VPg as well as C-terminal proteins P10 and P8 with ATPase and RNA binding properties, respectively. The processing of the P2a polyprotein takes place at consensus cleavage site E/T,S,N. Around 10% of ribosomes undergo −1 ribosomal frameshifting at the 3′ region of ORF2a and continue the translation on ORF2b. These ribosomes translate RNA-dependent RNA polymerase.
CP is translated from the subgenomic RNA (ORF3). The CP subunits are chemically identical but are not structurally equivalent. Three types of CP subunits termed A, B, C are related by quasi three-fold axes of symmetry and are involved in different inter-subunit contacts. The CP has two distinct domains. The N-terminal R (random) domain is localized to the interior of the particle. The S (shell) domain which forms the surface of the particle displays a canonical β-barrel motif. The arrangement of the N-terminal part of the subunit plays a crucial role in determining the capsid size. The CP of sobemoviruses has been reported to be necessary for cell-to-cell as well as for long-distance movement.
The molecular details of sobemovirus replication are not well understood. Recently, primer independent initiation of RNA synthesis by sesbania mosaic virus recombinant RNA-dependent RNA polymerase was demonstrated. For the minus-strand synthesis a stem-loop at the 3′ end of the genome is necessary. For the plus-strand synthesis, a conserved aCAAa motif at the 5′ end may be involved. The cellular compartments where the replication takes place are unknown.
Antigenic properties
Virions and coat proteins of sobemoviruses are efficient immunogens. A single precipitin line is formed in gel diffusion tests. There are distant serological relationships between some members of the genus. Several serotypes with different geographical origins have been identified in some species.
Biological properties
Host range
Sobemoviruses infect both dicotyledonous and monocotyledonous plants. However, the natural host range of each virus species is narrow except that sowbane mosaic virus (SoMV) can infect plants from several different families. Disease symptoms are mainly mosaic and mottle of infected leaves.
Transmission
All sobemoviruses are readily transmitted mechanically and many are also transmitted by beetles. In contrast, blueberry shoestring virus is transmitted by aphids and velvet tobacco mottle virus by mirids. SoMV has been reported to be transmitted by insects belonging to different orders (Diptera, Hemiptera, Thysanoptera), but this is probably non-specific mechanical transmission by the mouthparts of different insects. Several viruses in the genus are seed-transmissible.
Geographical distribution
There are members with limited distribution (one continent or one country only) but some species are found worldwide.
Cytopathic effects
Virions are found in both the cytoplasm and nuclei, and late in infection occur as large crystalline aggregates and inclusions in the cytoplasm and the vacuoles. Infected cells show cytoplasmic vacuolization. Occasionally the association of virus particles with chloroplast membranes and changes in chloroplast structure have been reported. Sobemoviruses have been found in mesophyll, epidermal as well as bundle sheath cells. In the course of systemic infection, viruses invade the phloem as well as the xylem and for some members virions are found in pit membranes of xylem cell walls.
Species demarcation criteria in the genus
- Genome sequence relatedness: different species have overall genome sequence identity less than about 75%
- Host range
- Serological relatedness may help in distinguishing species
List of species in the genus Sobemovirus
| Blueberry shoestring virus |
|
|
| Blueberry shoestring virus - USA |
| (BSSV-US) |
| Cocksfoot mottle virus |
|
|
| Cocksfoot mottle virus - Norway | [Z48630 = NC_002618] | (CfMV-NO) |
| Lucerne transient streak virus |
|
|
| Lucerne transient streak virus - New Zealand | [U31286 = NC_001696] | (LTSV-NZ) |
| Rice yellow mottle virus |
|
|
| Rice yellow mottle virus - Côte d'Ivoire: CI63 | [AJ608207] | (RYMV-CI63) |
| Ryegrass mottle virus |
|
|
| Ryegrass mottle virus - Japan | [EF091714] | (RGMoV-JP) |
| Sesbania mosaic virus |
|
|
| Sesbania mosaic virus - India | [AY004291 = NC_002568] | (SeMV-IN) |
| Solanum nodiflorum mottle virus |
|
|
| Solanum nodiflorum mottle virus - Australia |
| (SNMoV-AUS) |
| Southern bean mosaic virus |
|
|
| Southern bean mosaic virus - Brazil | [DQ875594] | (SBMV-BR) |
| Southern cowpea mosaic virus |
|
|
| Southern cowpea mosaic virus - USA | [M23021 = NC_001625] | (SCPMV-US) |
| Sowbane mosaic virus (Rubus chlorotic mottle virus) |
|
|
| Sowbane mosaic virus - USA | [GQ845002] | (SoMV-US) |
| Subterranean clover mottle virus |
|
|
| Subterranean clover mottle virus - Australia | [AY376453] | (SCMoV-MJ) |
| Turnip rosette virus |
|
|
| Turnip rosette virus - UK | [AY177608 = NC_004553] | (TRoV-UK) |
| Velvet tobacco mottle virus |
|
|
| Velvet tobacco mottle virus - Australia-K1 | [HM754263] | (VTMoV-K1) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Sobemovirus but have not been approved as species
| Cynosurus mottle virus |
| (CnMoV) |
| Ginger chlorotic fleck virus |
| (GCFV) |
| Imperata yellow mottle virus | [AM990928 = NC_011536] | (IYMV) |
| Papaya lethal yellowing virus | [GU066876*] | (PLYV) |
| Snake melon asteroid mosaic virus | [HM450304*] | (SMAMV) |
* Sequence does not comprise the complete genome.
Phylogenetic relationships within the genus
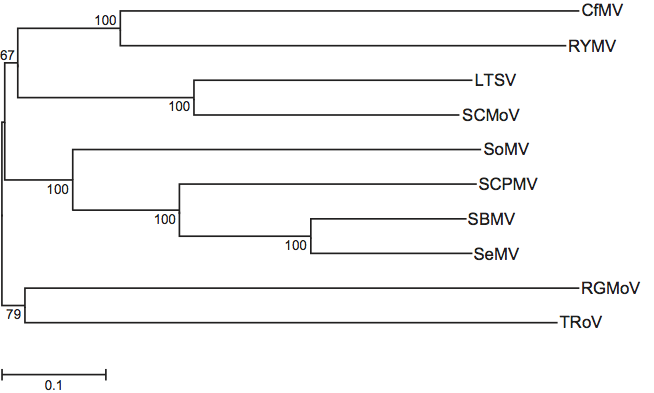
Phylogenetic relationships within the genus are depicted in Figure 3.
Similarity with other taxa
The 5′ ends of the sobemoviral genomes together with ORF1 are unrelated to any other known viral genera. However, the middle parts of the genomes (encoding the successive domains Pro-VPg-RdRP) are similar to those of the genera Polerovirus and Enamovirus from the family Luteoviridae. In contrast, the 3′ part of the sobemoviral genome encoding the CP is more closely related to CP genes of the genus Necrovirus of the family Tombusviridae. These similarities suggest early recombination events during the evolution of these genera. This possibility is further supported by the existence of Poinsettia latent virus, the unique species of the genus Polemovirus whose sequence shows a close relationship to poleroviruses within the first three quarters of its genome (encoding the P1 and polyprotein), but rather to sobemoviruses in the last quarter (encoding the CP gene). The unique species of the family Barnaviridae has a genomic organization similar to that of sobemoviruses (except that it lacks ORF1 of sobemoviruses) and its Pro-VPg-RdRP and CP genes are related to homologous sequences of sobemoviruses. In addition, the putative protease and RdRp of positive sense ss RNA animal viruses of the family Astroviridae have similarities to those of sobemoviruses.
Derivation of name
Sobemo: from the type species Southern bean mosaic virus.
Further reading
Albar, L., Bangratz-Reyser, M., Hébrard, E., Ndjiondjop, M., Jones, M. and Ghesquière, A. (2006). Mutations in the eIF(iso)4G translation initiation factor confer high resistance of rice to Rice yellow mottle virus. Plant J., 47, 417-426.
Fargette, D., Pinel, A., Abubakar, Z., Traoré, O., Brugidou, C., Fatogoma, S., Hébrard, E., Choisy, M., Séré, Y., Fauquet, C. and Konaté, G. (2004). Inferring the evolutionary history of Rice yellow mottle virus from genomic, phylogenetic and phylogeographic studies. J. Virol., 78, 3252-3261.
Gayathri, P., Satheshkumar, P., Prasad, K., Nair, S., Savithri, H. and Murthy, M. (2006). Crystal structure of the serine protease domain of Sesbania mosaic virus polyprotein and mutational analysis of residues forming the S1-binding pocket. Virology, 346, 440-451.
Govind, K. and Savithri, H. S. (2010). Primer independent initiation of RNA synthesis by SeMV recombinant RNA dependent RNA polymerase. Virology, 401, 280-292.
Lacombe, S., Bangratz, M., Vignols, F. and Brugidou, C. (2010). The rice yellow mottle virus P1 protein exhibits dual functions to suppress and activate gene silencing. Plant J., 61, 371-382.
Mäkeläinen, K. and Mäkinen, K. (2005). Factors affecting translation at the programmed -1 ribosomal frameshifting site of Cocksfoot mottle virus RNA in vivo. Nucleic Acids Res., 33, 2239-2247.
Meier, M. and Truve, E. (2007). Sobemoviruses possess a common CfMV-like genomic organization. Arch. Virol., 152, 635-640.
Nair, S. and Savithri, H. (2010). Processing of SeMV polyproteins revisited. Virology, 396, 106-117.
Nair, S. and Savithri, H.S. (2010). Natively unfolded nucleic acid binding P8 domain of SeMV polyprotein 2a affects the novel ATPase activity of the preceding P10 domain. FEBS Lett., 584, 571-576.
Qu, C., Liljas, L., Opalka N., Brugidou, C., Yeager, M., Beachy, R., Fauquet, C., Johnson, J. and Lin, T. (2000). 3D domain swapping of a molecular switch for quasi-equivalent symmetry modulates the stability of an icosahedral virus group. Structure, 8, 1095-1103.
Contributed by
Truve, E. and Fargette, D.
Figures
Figure 1 (Left) Electronic rendering of particles of southern bean mosaic virus (T=3); (centre) diagrammatic representation of a T=3 structure capsid; (right) negative contrast electron micrograph of rice yellow mottle virus particles stained in uranyl acetate. The bar represents 100 nm.
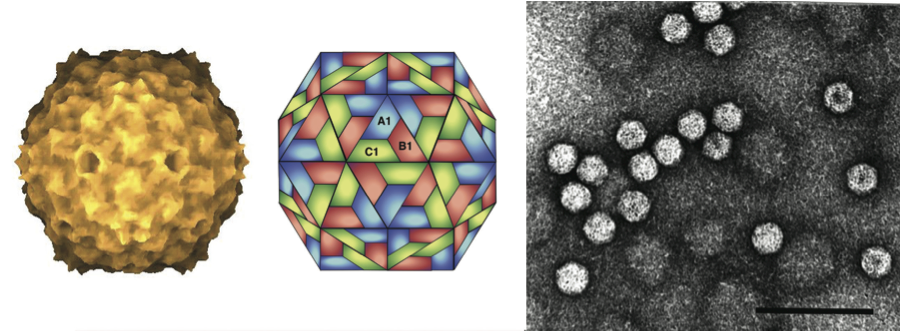
Figure 2 Diagram of the genome organization of southern bean mosaic virus (SBMV).
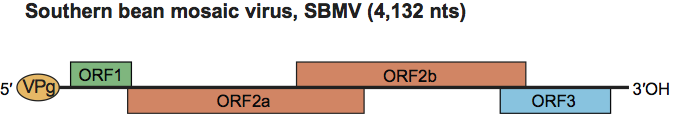
Figure 3 Phylogenetic tree inferred by the maximum composite likelihood model from the full sequences of members of the genus Sobemovirus. Mid-point rooting was applied and the percentage of bootstrap support of the branches shown at the nodes.