Family: Potyviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Distinguishing features
The family Potyviridae consists of plant viruses with a single stranded, positive sense RNA genome and flexuous, filamentous particles. Genomes have a VPg covalently linked to the 5′ end and the 3′ terminus is polyadenylated. Genomes encode a large polyprotein that is self-cleaved into a set of functional proteins. Gene order and protein sequences are conserved throughout the family.
Virion properties
Morphology
Virions are flexuous filaments with no envelope and are 11–15 nm in diameter, with a helical pitch of about 3.4 nm (Figure 1). Particle lengths of members of some of the six genera differ. Members of the genera Potyvirus, Ipomovirus, Macluravirus, Rymovirus, Tritimovirus, Brambyvirus and the unassigned viruses are monopartite with particle modal lengths of 650–900 nm; members of the genus Bymovirus are bipartite with particles of two modal lengths of 250–300 and 500–600 nm.
Physicochemical and physical properties
Virions of viruses in the genera Potyvirus and Rymovirus have a density in CsCl of about 1.31 g cm−3 and S20,w of 137–160S. Those of the genus Bymovirus have a density in CsCl of about 1.29 g cm−3.
Nucleic acid
Viruses in all genera except Bymovirus have a single molecule of positive sense, ssRNA, 9.3–10.8 kb in size. Virions are infectious. A VPg of about 24 kDa is covalently linked to the 5′-terminal nt. A polyadenylate tract (20 to 160 adenosines) is present at the 3′ terminus. Bymoviruses have two positive sense, ssRNA molecules; RNA-1 is 7.3–7.6 kb in size and RNA-2 is 3.5–3.7 kb in size. Both RNAs have 3′-terminal polyadenylate tracts and probably a VPg at the 5′ termini.
Proteins
Virions contain one type of CP of 28.5–47 kDa. N- and C-terminal residues are positioned on the exterior of the virion. Mild trypsin treatment removes N- and C-terminal segments, leaving a trypsin-resistant core of about 24 kDa. Plant proteases may degrade the CP in vivo, as occurs in vitro during purification using some procedures or from certain hosts. All potyvirus CPs display significant aa sequence identity in the trypsin-resistant core, but little identity in their N and C-terminal segments.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genomic RNA (each genomic RNA for the genus Bymovirus) encodes a single major polyprotein. This then undergoes co- and post-translational proteolytic processing by three viral-encoded proteinases to form individual gene products. Genomic RNA replicates via the production of a full-length negative sense RNA. While there are exceptions noted in the relevant genus descriptions, the polyprotein of the majority of monopartite viruses in the family is cleaved into ten products which show conservation of sequence and organisation. As shown in Figure 2, these products are:
P1: Of all the potyvirid proteins, P1 is the least conserved in sequence and the most variable in size. It plays a significant role in virus replication probably due to the stimulation of the gene silencing suppressor HC-Pro. A serine protease domain towards the C-terminus cleaves the P1 from the polyprotein, typically at Tyr/Phe-Ser.
HC-Pro (Helper Component-Protease): the HC-Pro protein has roles in suppression of gene silencing and in vector transmission. A cysteine protease domain towards the C-terminus cleaves it from the remainder of the downstream polyprotein, typically at Gly-Gly.
P3: Involved in virus replication and appears to be significant in host range and symptom development.
6K1: The function of this small protein is not known.
CI (Cylindrical Inclusion protein): This protein has helicase activity and accumulates in inclusion bodies in the cytoplasm of infected plant cells.
6K2: A small transmembrane protein probably anchoring the replication complex to the ER.
VPg (Viral Protein genome-linked): Attached to the 5′ terminus of the genome and belongs to a class of intrinsically disordered proteins. It plays multiple roles in the viral infection cycle. It is essential for virus replication and translation, interacting with one or several isoforms of the eIF4E translation initiation factor. It is involved in suppression of RNA silencing.
NIa-Pro: Serine-like cysteine protease responsible for cleavage of most sites in the polyprotein, typically at Gln/Glu-(Ser/Gly/Ala).
NIb: The RNA-dependent RNA polymerase.
CP: Viral coat protein that also has roles in virus movement, genome amplification and vector transmission.
Recent studies have shown the presence of an additional short ORF (PIPO=“pretty interesting potyvirus ORF”) embedded within the P3 cistron and expressed as a P3_PIPO fusion product via ribosomal frameshifting. This has now been identified throughout the family and has been shown to be essential for virus intercellular movement.
Antigenic properties
The viral proteins are moderately immunogenic; there are serological relationships among members. An epitope of the CP in the conserved internal trypsin-resistant core has been identified that is similar in most members of the family.
Biological properties
Inclusion body formation
All members of the family Potyviridae form cytoplasmic cylindrical inclusion (CI) bodies during infection. The CI is an array of a 70 kDa viral protein that possesses ATPase and helicase activities. Some potyviruses induce nuclear inclusion bodies that are co-crystals of two viral-encoded proteins – NIa and NIb – present in equimolar amounts. The small nuclear inclusion (NIa) protein (49 kDa) is a polyprotein consisting of the VPg and proteinase. Amorphous inclusion bodies are also evident in the cytoplasm during certain potyvirus infections and represent aggregations of the protein HC-Pro and perhaps other non-structural proteins.
Host range
Some members have a narrow host range, most members infect an intermediate number of plants, and a few members infect species in up to 30 families. Transmission to most hosts is readily accomplished by mechanical inoculation. Many viruses are widely distributed. Distribution is aided by seed transmission in some cases.
Transmission
Potyvirids are vectored by a variety of organisms. Members of the genera Potyvirus and Macluravirus have aphid vectors that transmit in a non-persistent, non-circulative manner. A helper component and a particular CP aa triplet (i.e., DAG for some potyviruses) are required for aphid transmission. Rymoviruses and tritimoviruses are transmitted by eriophyid mites, in a semi-persistent manner. Bymoviruses are transmitted by root-infecting vectors in the order Plasmodiophorales, once described as fungi but now classified as Cercozoa. Ipomoviruses appear to be transmitted by whiteflies.
Species demarcation criteria
Throughout the family, species are distinguished by the following criteria:
- Genome sequence relatedness: different species have CP aa sequence identity less than about 80%; and nt sequence identity less than 76% either in the CP or over the whole genome. There are also differences in polyprotein cleavage sites.
- Host range and key host reactions; lack of cross protection.
- Different inclusion body morphology.
- Antigenic properties: serological relatedness may help in distinguishing species.
Genus Potyvirus
Type species Potato virus Y
Distinguishing features
The largest genus in the family contains viruses transmitted by aphids in a non-persistent manner.
Virion properties
Morphology
Virions are flexuous filaments, 680–900 nm long and 11–13 nm wide, with helical symmetry and a pitch of about 3.4 nm. Particles of some viruses are longer in the presence of divalent cations than in the presence of EDTA.
Physicochemical and physical properties
Virion S20,w is 137–160S; density in CsCl is 1.31 g cm−3; E0.1%1 cm, 260 nm=2.4–2.7.
Nucleic acid
Virions contain a single molecule of linear, positive sense ssRNA, about 9.7 kb in size; virions contain 5% RNA by weight.
Proteins
Virions contain a single CP, 30–47 kDa in size. The CP of most isolates of the type species, PVY, contains 267 aa.
Genome organization and replication
The genome is organized as described earlier (Figure 2).
Antigenic properties
Virions are moderately immunogenic; there are serological relationships among many members. Some monoclonal antibodies react with most aphid-transmitted potyviruses. The CP aa sequence identity among aphid-transmitted viruses is 40–70%. Some viruses are serologically related to viruses in the genera Rymovirus and Bymovirus.
Biological properties
Many individual viruses have a narrow host range, but a few infect plant species in up to 30 host families. The viruses are transmitted by aphids in a non-persistent manner and are transmissible experimentally by mechanical inoculation. Some isolates are inefficiently transmitted by aphids and others are not transmissible by aphids at all. This is apparently due to mutations within the helper component and/or CP cistrons. Some viruses are seed-transmitted.
List of species in the genus Potyvirus
| Algerian watermelon mosaic virus |
|
|
| Algerian watermelon mosaic virus-Algeria: H4 | [EU410442=NC_010736] | (AWMV-H4) |
| Alstroemeria mosaic virus |
|
|
| Alstroemeria mosaic virus-O1 | [AB158522*] | (AlMV-O1) |
| Alternanthera mild mosaic virus |
|
|
| Alternanthera mild mosaic virus-Brazil | [EF442668*] | (AltMMV-BR) |
| Amaranthus leaf mottle virus |
|
|
| Amaranthus leaf mottle virus-Italy | [AJ580095*] | (AmLMV-I) |
| Amazon lily mosaic virus |
|
|
| Amazon lily mosaic virus-Japan | [AB158523*] | (ALiMV-JP) |
| Angelica virus Y |
|
|
| Angelica virus Y-USA:g | [EF488741*] | (AVY-g) |
| Apium virus Y |
|
|
| Apium virus Y-USA: Ce | [HM363516=NC_014905] | (ApVY-Ce) |
| Araujia mosaic virus |
|
|
| Araujia mosaic virus-Argentina: ARG1973 | [EF710625*] | (ArjMV-ARG1973) |
| Arracacha mottle virus |
|
|
| Arracacha mottle virus-Brazil:C17 | [DQ925486*] | (AMoV-C17) |
| Artichoke latent virus |
|
|
| Artichoke latent virus-California |
| (ArLV-Cal) |
| Asparagus virus 1 |
|
|
| Asparagus virus 1-Germany |
| (AV1-DE) |
| Banana bract mosaic virus |
|
|
| Banana bract mosaic virus-Philippines | [DQ851496=NC_009745] | (BBrMV-PH) |
| Basella rugose mosaic virus |
|
|
| Basella rugose mosaic virus-Taiwan:AC | [DQ821938=NC_009741] | (BaRMV-AC) |
| Peace lily mosaic virus | [DQ851494] | (PcLMV) |
| Bean common mosaic necrosis virus |
|
|
| Bean common mosaic necrosis virus-USA:NL-3 | [AY282577] | (BCMNV-NL3) |
| Bean common mosaic virus |
|
|
| Bean common mosaic virus-NL4 | [DQ666332] | (BCMV-NL4) |
| Azuki bean mosaic virus | [AB012663*] | (AzBMV) |
| Blackeye cowpea mosaic virus | [AJ312437=NC_003397] | (BlCMV) |
| Dendrobium mosaic virus | [U23564*] | (DeMV) |
| Peanut stripe virus | [U34972] | (PStV) |
| Yam bean mosaic virus | [AB289438*] | (YBMV) |
| Bean yellow mosaic virus |
|
|
| Bean yellow mosaic virus-MB4 | [D83749=NC_003492] | (BYMV-MB4) |
| Beet mosaic virus |
|
|
| Beet mosaic virus-Wa | [AY206394=NC_005304] | (BtMV-Wa) |
| Bidens mottle virus |
|
|
| Bidens mottle virus-Taiwan:B12 | [EU250210] | (BiMoV-B12) |
| Sunflower chlorotic spot virus | [AF538686*] | (SuCSV) |
| Brugmansia suaveolens mottle virus |
|
|
| Brugmansia suaveolens mottle virus-Brazil | [AB551370=NC_014536] | (BsMoV-BR) |
| Butterfly flower mosaic virus |
|
|
| Butterfly flower mosaic virus-China:HZ | [AM774001*] | (BFMV-HZ) |
| Calanthe mild mosaic virus |
|
|
| Calanthe mild mosaic virus-Japan | [AB011404*] | (CalMMV-JP) |
| Canna yellow streak virus |
|
|
| Canna yellow streak virus-UK | [GQ421689=NC_013261] | (CaYSV-UK) |
| Carnation vein mottle virus |
|
|
| Carnation vein mottle virus-Japan | [AB017630*] | (CVMoV-JP) |
| Carrot thin leaf virus |
|
|
| Carrot thin leaf virus-Australia | [AF203530*] | (CTLV-AU) |
| Carrot virus Y |
|
|
| Carrot virus Y-Australia:Victoria | [AF203537*] | (CarVY-VI) |
| Celery mosaic virus |
|
|
| Celery mosaic virus-Netherlands | [AF203531*] | (CeMV-NL) |
| Ceratobium mosaic virus |
|
|
| Ceratobium mosaic virus-Australia:13 | [AF022442*] | (CerMV-13) |
| Chilli ringspot virus |
|
|
| Chilli ringspot virus-Vietnam:C8 | [DQ925438*] | (ChiRSV-C8) |
| Chilli veinal mottle virus |
|
|
| Chilli veinal mottle virus-Pepper vein banding virus | [AJ237843=NC_005778] | (ChiVMV-PVB) |
| Chinese artichoke mosaic virus |
|
|
| Chinese artichoke mosaic virus-Japan | [AB099711*] | (ChAMV-JP) |
| Clitoria virus Y |
|
|
| Clitoria virus Y-Australia:Queensland | [AF228515*] | (ClVY-QD) |
| Clover yellow vein virus |
|
|
| Clover yellow vein virus-30 | [AB011819=NC_003536] | (ClYVV-30) |
| Cocksfoot streak virus |
|
|
| Cocksfoot streak virus-Germany | [AF499738= NC_003742] | (CSV-DE) |
| Colombian datura virus |
|
|
| Colombian datura virus-Germany:Br1 | [AJ237921*] | (CDV-Br1) |
| Petunia flower mottle virus | [AF030689*] | (PFMoV) |
| Commelina mosaic virus |
|
|
| Commelina mosaic virus-Florida |
| (ComMV-FL) |
| Cowpea aphid-borne mosaic virus |
|
|
| Cowpea aphid-borne mosaic virus-Zimbabwe | [AF348210=NC_004013] | (CABMV-ZM) |
| Cowpea green vein banding virus |
|
|
| Cowpea green vein banding virus-Brazil |
| (CGVBV-BR) |
| Cypripedium virus Y |
|
|
| Cypripedium virus Y-UK:CP | [AF185954*] | (CypVY-CP) |
| Daphne mosaic virus |
|
|
| Daphne mosaic virus-Czech Republic | [DQ299908=NC_008028] | (DapMV-CZ) |
| Dasheen mosaic virus |
|
|
| Dasheen mosaic virus-China: M13 | [AJ298033=NC_003537] | (DsMV-M13) |
| Vanilla mosaic virus | [AJ616719*] | (VanMV) |
| Datura shoestring virus |
|
|
| Datura shoestring virus-India:Simla |
| (DSSV-IN) |
| Diuris virus Y |
|
|
| Diuris virus Y-Australia | [AF203527*] | (DiVY-AU) |
| East Asian passiflora virus |
|
|
| East Asian passiflora virus-Japan:AO | [AB246773=NC_007728] | (EAPV-AO) |
| Endive necrotic mosaic virus |
|
|
| Endive necrotic mosaic virus-Germany 1/85 | [AJ223827*] | (ENMV-1/85) |
| Euphorbia ringspot virus |
|
|
| Euphorbia ringspot virus-USA | [AY697300*] | (EuRSV-US) |
| Freesia mosaic virus |
|
|
| Freesia mosaic virus-South Korea | [GU214748] | (FreMV-KO) |
| Fritillary virus Y |
|
|
| Fritillary virus Y-China:Pan’an | [AM039800=NC_010954] | (FVY-PA) |
| Gloriosa stripe mosaic virus |
|
|
| Gloriosa stripe mosaic virus-Netherlands | [EU042761*] | (GSMV-NL) |
| Groundnut eyespot virus |
|
|
| Groundnut eyespot virus-Ivory Coast |
| (GEV-IC) |
| Guinea grass mosaic virus |
|
|
| Guinea grass mosaic virus-Ivory Coast |
| (GGMV-IC) |
| Hardenbergia mosaic virus |
|
|
| Hardenbergia mosaic virus-Australia: BB-6 | [DQ898188*] | (HaMV-BB-6) |
| Helenium virus Y |
|
|
| Helenium virus Y-Germany |
| (HVY-DE) |
| Henbane mosaic virus |
|
|
| Henbane mosaic virus-Hungary:PHYS/H | [AM184113*] | (HMV-PHYS/H) |
| Hibbertia virus Y |
|
|
| Hibbertia virus Y-Australia: New South Wales | [AF228516*] | (HiVY-NSW) |
| Hippeastrum mosaic virus |
|
|
| Hippeastrum mosaic virus-Amaryllis | [AY566239*] | (HiMV-AM) |
| Hyacinth mosaic virus |
|
|
| Hyacinth mosaic virus-Netherlands | [EF203681*] | (HyaMV-NL) |
| Iris fulva mosaic virus |
|
|
| Iris fulva mosaic virus-USA:Massachusetts |
| (IFMV-US) |
| Iris mild mosaic virus |
|
|
| Iris mild mosaic virus-New Zealand:DC4a | [DQ436918*] | (IMMV-DC4a) |
| Iris severe mosaic virus |
|
|
| Iris severe mosaic virus-Netherlands | [X75939*] | (ISMV-NL) |
| Japanese yam mosaic virus |
|
|
| Japanese yam mosaic virus-mild | [AB027007=NC_000947] | (JYMV-mild) |
| Johnsongrass mosaic virus |
|
|
| Johnsongrass mosaic virus-Australia | [Z26920=NC_003606] | (JGMV-AU) |
| Kalanchoë mosaic virus |
|
|
| Kalanchoë mosaic virus-Netherlands:F39 | [GQ497731*] | (KMV-F39) |
| Konjac mosaic virus |
|
|
| Konjac mosaic virus-Japan: F | [AB219545=NC_007913] | (KoMV-F) |
| Zantedeschia mosaic virus | [AF470620*] | (ZaMV) |
| Japanese hornwort mosaic virus | [AB081518*] | (JHMV) |
| Leek yellow stripe virus |
|
|
| Leek yellow stripe virus-China:Yuhang | [AJ307057=NC_004011] | (LYSV-YH) |
| Lettuce mosaic virus |
|
|
| Lettuce mosaic virus-E | [X97705=NC_003605] | (LMV-E) |
| Lily mottle virus |
|
|
| Lily mottle virus-China:Sb | [AJ564636=NC_005288] | (LMoV-Sb) |
| Lycoris mild mottle virus |
|
|
| Lycoris mild mottle virus-Taiwan | [AF399672*] | (LyMMoV-TW) |
| Maize dwarf mosaic virus |
|
|
| Maize dwarf mosaic virus-Bulgaria | [AJ001691=NC_003377] | (MDMV-BU) |
| Malva vein clearing virus |
|
|
| Malva vein clearing virus-Italy:DS-Ba-01 | [FM212972*] | (MVCV-DS-Ba-01) |
| Meadow saffron breaking virus |
|
|
| Meadow saffron breaking virus-France | [AY388995*] | (MSBV-FR) |
| Moroccan watermelon mosaic virus |
|
|
| Moroccan watermelon mosaic virus-Tunisia:TN05-76 | [EF579955=NC_009995*] | (MWMV-TN05-76) |
| Narcissus degeneration virus |
|
|
| Narcissus degeneration virus-China:Zhangzhou | [AM182028=NC_008824] | (NDV-ZZ) |
| Narcissus late season yellows virus |
|
|
| Narcissus late season yellows virus-China:Hangzhou 2 | [AJ493579*] | (NLSYV-HZ2) |
| Narcissus yellow stripe virus |
|
|
| Narcissus yellow stripe virus-China:Zhangzhou | [AM158908=NC_011541] | (NYSV-ZZ) |
| Nerine yellow stripe virus |
|
|
| Nerine yellow stripe virus-Netherlands:Ne800 | [EF362621*] | (NeYSV-Ne800) |
| Nothoscordum mosaic virus |
|
|
| Nothoscordum mosaic virus-Canary Islands |
| (NoMV-CAY) |
| Onion yellow dwarf virus |
|
|
| Onion yellow dwarf virus-China:Yuhang | [AJ510223=NC_005029] | (OYDV-YH) |
| Ornithogalum mosaic virus |
|
|
| Ornithogalum mosaic virus-O | [D00615*] | (OrMV-O) |
| Ornithogalum virus 2 |
|
|
| Ornithogalum virus 2-Japan:Akita, Oga | [AB271783*] | (OrV2-AO) |
| Ornithogalum virus 3 |
|
|
| Ornithogalum virus 3-Japan | [AB282754*] | (OrV3-JP) |
| Papaya leaf distortion mosaic virus |
|
|
| Papaya leaf distortion mosaic virus-Japan:P | [AB088221] | (PLDMV-P) |
| Papaya ringspot virus |
|
|
| Papaya ringspot virus-Hawaii | [S46722=X67673] | (PRSV-HAT ) |
| Parsnip mosaic virus |
|
|
| Parsnip mosaic virus-UK:Scotland |
| (ParMV-UK) |
| Passiflora chlorosis virus |
|
|
| Passiflora chlorosis virus-LAJ-2006 | [DQ860147*] | (PaChV-Pangda15) |
| Passion fruit woodiness virus |
|
|
| Passion fruit woodiness virus-Australia:MU2 | [HQ122652=NC_014790] | (PWV-MU2) |
| Pea seed-borne mosaic virus |
|
|
| Pea seed-borne mosaic virus-DPD1 | [D10930=NC_001671] | (PSbMV-DPD1) |
| Peanut mottle virus |
|
|
| Peanut mottle virus-M | [AF023848=NC_002600] | (PeMoV-M) |
| Pennisetum mosaic virus |
|
|
| Pennisetum mosaic virus-China:B | [AY642590=NC_007147] | (PenMV-B) |
| Pepper mottle virus |
|
|
| Pepper mottle virus-California | [M96425=NC_001517] | (PepMoV-Cal) |
| Pepper severe mosaic virus |
|
|
| Pepper severe mosaic virus-South Korea | [AM181350=NC_008393] | (PepSMV-KO) |
| Pepper veinal mottle virus |
|
|
| Pepper veinal mottle virus-P | [DQ645484=NC_011918] | (PVMV-P) |
| Pepper yellow mosaic virus |
|
|
| Pepper yellow mosaic virus-Brazil:Pi-15 | [AB541985=NC_014327] | (PepYMV-Pi15) |
| Peru tomato mosaic virus |
|
|
| Peru tomato mosaic virus-Peru:PPK13 | [AJ437280=NC_004573] | (PTV-PPK13) |
| Pfaffia mosaic virus |
|
|
| Pfaffia mosaic virus-Brazil | [AY485276*] | (PfMV-BR) |
| Pleione virus Y |
|
|
| Pleione virus Y-Australia | [AF185958*] | (PlVY-AU) |
| Plum pox virus |
|
|
| Plum pox virus-NAT | [D13751=NC_001445] | (PPV-NAT) |
| Pokeweed mosaic virus |
|
|
| Pokeweed mosaic virus-USA |
| (PkMV-USA) |
| Potato virus A |
|
|
| Potato virus A-Hungary: B11 | [AJ296311=NC_004039] | (PVA-B11) |
| Tamarillo mosaic virus | [AJ131403] | (TamMV) |
| Potato virus V |
|
|
| Potato virus V-UK:DV 42 | [AJ243766=NC_004010] | (PVV-DV42) |
| Potato virus Y |
|
|
| Potato virus Y-France:O | [X12456=NC_001616] | (PVY-O) |
| Potato virus Y-Hungary:N | [M95491] | (PVY-N) |
| Potato virus Y-France:C | [AJ890348] | (PVY-C) |
| Bidens mosaic virus | [AY960150*] | (BiMV) |
| Ranunculus leaf distortion virus |
|
|
| Ranunculus leaf distortion virus-Italy:RN122 | [DQ152190*] | (RanLDV-RN122) |
| Ranunculus mild mosaic virus |
|
|
| Ranunculus mild mosaic virus-Italy:RN129 | [DQ152191*] | (RanMMV-RN129) |
| Ranunculus mosaic virus |
|
|
| Ranunculus mosaic virus-Italy:RN136 | [DQ152192*] | (RanMV-RN136) |
| Rhopalanthe virus Y |
|
|
| Rhopalanthe virus Y-Australia | [AF185956*] | (RhVY-AU) |
| Sarcochilus virus Y |
|
|
| Sarcochilus virus Y-Australia | [AF185957*] | (SaVY-AU) |
| Scallion mosaic virus |
|
|
| Scallion mosaic virus-China:Hangzhou | [AJ316084=NC_003399] | (ScaMV-HZ) |
| Shallot yellow stripe virus |
|
|
| Shallot yellow stripe virus-China:ZQ2 | [AJ865076=NC_007433] | (SYSV-ZQ2) |
| Sorghum mosaic virus |
|
|
| Sorghum mosaic virus-China:Xiaoshan | [AJ310197=NC_004035] | (SrMV-Xiaoshan) |
| Soybean mosaic virus |
|
|
| Soybean mosaic virus-N | [D00507=NC_002634] | (SMV-N) |
| Spiranthes mosaic virus 3 |
|
|
| Spiranthes mosaic virus 3-USA | [AY685218*] | (SpMV3-USA) |
| Sugarcane mosaic virus |
|
|
| Sugarcane mosaic virus-China:Hangzhou | [AJ297628=NC_003398] | (SCMV-HZ) |
| Sunflower mosaic virus |
|
|
| Sunflower mosaic virus-USA:Texas | [AF465545*] | (SuMV-TX) |
| Sweet potato feathery mottle virus |
|
|
| Sweet potato feathery mottle virus-S | [D86371=NC_001841] | (SPFMV-S) |
| Sweet potato latent virus |
|
|
| Sweet potato latent virus-Taiwan | [X84012*] | (SPLV-TW) |
| Sweet potato mild speckling virus |
|
|
| Sweet potato mild speckling virus-Argentina | [U61228*] | (SPMSV-AR) |
| Sweet potato virus 2 |
|
|
| Sweet potato virus 2-Nigeria | [AY232437*] | (SPV2-NG) |
| Sweet potato virus G |
|
|
| Sweet potato virus G-Peru:Hua2 | [EU218528*] | (SPVG-Hua2) |
| Telfairia mosaic virus |
|
|
| Telfairia mosaic virus-Nigeria |
| (TeMV-NI) |
| Telosma mosaic virus |
|
|
| Telosma mosaic virus-Vietnam:Hanoi | [DQ851493=NC_009742] | (TelMV-VN) |
| Thunberg fritillary mosaic virus |
|
|
| Thunberg fritillary mosaic virus-China: Ningbo | [AJ851866=NC_007180] | (TFMV-NB) |
| Tobacco etch virus |
|
|
| Tobacco etch virus-HAT | [M11458=NC_001555] | (TEV-HAT ) |
| Tobacco vein banding mosaic virus |
|
|
| Tobacco vein banding mosaic virus-China:YND | [EF219408=NC_009994] | (TVBMV-YND) |
| Tobacco vein mottling virus |
|
|
| Tobacco vein mottling virus-S | [U38621] | (TVMV-S) |
| Tradescantia mild mosaic virus |
|
|
| Tradescantia mild mosaic virus-Italy:IFA195 | [AY861351*] | (TraMMV-IFA195) |
| Tropaeolum mosaic virus |
|
|
| Tropaeolum mosaic virus-Ecuador:Mashua |
| (TrMV-EC) |
| Tuberose mild mosaic virus |
|
|
| Tuberose mild mosaic virus-Taiwan | [AF062926*] | (TuMMV-TW) |
| Tuberose mild mottle virus |
|
|
| Tuberose mild mottle virus-China:Hangzhou | [AJ581528*] | (TuMMoV-HZ) |
| Tulip breaking virus |
|
|
| Tulip breaking virus-India |
| (TBV-IN) |
| Tulip mosaic virus |
|
|
| Tulip mosaic virus-Japan | [X63630*] | (TulMV-JP) |
| Turnip mosaic virus |
|
|
| Turnip mosaic virus-UK1 | [AF169561=NC_002509] | (TuMV-UK1) |
| Twisted-stalk chlorotic streak virus |
|
|
| Twisted-stalk chlorotic streak virus-Alaska:Denali 2001 | [AY954248*] | (TSCSV-Denali 2001) |
| Vallota mosaic virus |
|
|
| Vallota mosaic virus-USA:Beltsville | [EF441726*] | (ValMV-BV) |
| Watermelon leaf mottle virus |
|
|
| Watermelon leaf mottle virus-Florida | [AF028004*] | (WLMV-FL) |
| Watermelon mosaic virus |
|
|
| Watermelon mosaic virus-Fr | [AY437609=NC_006262] | (WMV-Fr) |
| Wild potato mosaic virus |
|
|
| Wild potato mosaic virus-Peru | [AJ437279=NC_004426] | (WPMV-Peru) |
| Wild tomato mosaic virus |
|
|
| Wild tomato mosaic virus-Vietnam: Laichau | [DQ851495=NC_009744] | (WTMV-VN) |
| Wisteria vein mosaic virus |
|
|
| Wisteria vein mosaic virus-China:Beijing | [AY656816=NC_007216] | (WVMV-BJ) |
| Yam mild mosaic virus |
|
|
| Yam mild mosaic virus-Papua New Guinea | [AB022424*] | (YMMV-PNG) |
| Yam mosaic virus |
|
|
| Yam mosaic virus-Ivory Coast | [U42596=NC_004752] | (YMV-IC) |
| Zantedeschia mild mosaic virus |
|
|
| Zantedeschia mild mosaic virus-Taiwan | [AY626825=NC_011560] | (ZaMMV-TW) |
| Zea mosaic virus |
|
|
| Zea mosaic virus-Israel | [AF228693*] | (ZeMV-IS) |
| Zucchini yellow fleck virus |
|
|
| Zucchini yellow fleck virus-Italy | [DQ641510*] | (ZYFV-IT) |
| Zucchini yellow mosaic virus |
|
|
| Zucchini yellow mosaic virus-Taiwan:TN3 | [AF127929=NC_003224] | (ZYMV-TN3 ) |
Species names are in italic script; names of strains, isolates and synonyms are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Partial sequence including the coat protein; complete genome sequence not available.
List of other related viruses which may be members of the genus Potyvirus but have not been approved as species
| Ammi majus latent virus | [AB361564*] | (AmLV) |
| Anemone mosaic virus | [EU042755*] | (AnMV) |
| Arisaema potyvirus 1 | [FJ546415*] | (APV1) |
| Arisaema potyvirus 2 | [FJ546416*] | (APV2) |
| Begonia flower breaking virus | [FJ539085*] | (BFBV) |
| Calla lily latent virus | [EF105297] | (CLLV) |
| Catharanthus mosaic virus | [DQ365928*] | (CatMV) |
| Chickpea yellow mosaic virus | [AF527879*] | (CpYMV) |
| Christmas bell potyvirus | [EF427894] | (CBPV) |
| Delphinium vein-clearing virus | [FJ349327*] | (DVCV) |
| Ecuadorian rocoto virus | [EU495234*] | (EcRV) |
| Impatiens flower break virus | [AY864851*] | (IFBV) |
| Lupin mosaic virus | [EU847625=NC_014898] | (LuMV) |
| Muscari mosaic virus | [EU042752*] | (MuMV) |
| Omphalodes virus Y | [AY974328*] | (OmVY) |
| Ornamental onion stripe mosaic virus | [EU042750*] | (OOSMV) |
| Ornithogalum necrotic mosaic virus | [FJ159380*] | (OrNMV) |
| Ornithogalum stripe mosaic virus | [FJ159376*] | (OrSMV) |
| Ornithogalum virus 4 | [EU042753*] | (OrV4) |
| Panax virus Y | [GQ916624= NC_014252] | (PnVY) |
| Passiflora foetida virus Y | [DQ112219*] | (PfVY) |
| Siratro 1 virus Y | [DQ098900*] | (SVY1) |
| Siratro 2 virus Y | [DQ098901*] | (SVY2) |
| Snowdrop virus Y | [EU927399*] | (SnVY) |
| Sunflower chlorotic mottle virus | [GU181199=NC_014038] | (SCMoV) |
| Stenomesson mosaic virus | [EU042757*] | (StMV) |
| Sweet potato virus C | [AB509453] | (SPVC) |
| Trillium crinkled leaf virus | [FJ648825*] | (TCLV) |
| Vanilla distortion mosaic virus | [AY943944*] | (VDMV) |
| Veltheimia mosaic virus | [EF203686*] | (VelMV) |
| Veltheimia virus Y | [EU684971*] | (VelVY) |
| Verbena virus Y | [EU564817=NC_010735] | (VerVY) |
* Partial sequence including the coat protein; complete genome sequence not available.
Genus Brambyvirus
Type species Blackberry virus Y
Distinguishing features
This is a monotypic genus. The single species is distinguished from all other members of the family by having a very large P1 protein (83.6 k Da) containing an AlkB domain. It is also phylogenetically distinct.
Virion properties
Morphology
Virions are flexuous filaments 800×11–15 nm in size.
Physicochemical and physical properties
No information.
Nucleic acid
Virions contain a single molecule of linear positive sense ssRNA with a 3′-poly(A) terminus. Virion RNA is about 11 kb in size.
Proteins
There is a single CP of 40.9 kDa.
Genome organization and replication
Apart from the size of the P1, the genome organization is identical to that of most monopartite viruses in the family Potyviridae (Figure 2).
Antigenic properties
The virus could not be detected by a universal potyvirus monoclonal antibody but there are no additional data.
Biological properties
Host range
The virus has been reported only from wild and cultivated blackberry (Rubus sp.) where it is often symptomless but is also a component of a complex of viruses. It is not known to cause symptoms in any herbaceous test host.
Transmission
The virus is presumed to be transmitted by an aerial vector that has not yet been identified.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Brambyvirus
| Blackberry virus Y |
|
|
| Blackberry virus Y-Arkansas 3 | [AY994084=NC_008558] | (BlVY-Ark3) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Brambyvirus but have not been approved as species
None reported.
Genus Ipomovirus
Type species Sweet potato mild mottle virus
Distinguishing features
Ipomoviruses are distinguished from other genera by their mode of transmission (whiteflies) and by phylogenetic analyses.
Virion properties
Morphology
Virions are flexuous filaments 800–950 nm long.
Physicochemical and physical properties
Virion S20,w is 155S for sweet potato mild mottle virus (SPMMV).
Nucleic acid
Virions contain a positive sense ssRNA, with a 3′-poly(A) terminus.
Proteins
The viral CP is a single polypeptide of 302–378 aa (35–41 kDa).
Genome organization and replication
Ipomovirus genomes consist of 9069–10818 nt excluding the 3′-terminal poly(A) tail and encode a polyprotein of 2902–3456 aa (Figure 3). These viruses exhibit unusual structural variability. The structure and organization of the SPMMV genome is similar to Potyvirus, but some motifs of HC-Pro and CP characteristic of Potyvirus are incomplete or missing, which may account for its vector relations. The unusually large P1 (83 kDa) of SPMMV contains no obvious AlkB domain and hence differs from Brambyvirus. Cucumber vein yellowing and squash vein yellowing viruses (CVYV and SqVYV) differ from SPMMV by containing two P1-like serine proteases (P1a and P1b) but no HC-Pro. P1b functions as a suppressor of RNA silencing. Cassava brown streak virus (CBSV) differs from SPMMV by having no HC-Pro and, also, from CVYV and SqVYV by having only P1b which suppresses silencing. Additionally, CBSV contains a Maf/HAM1-like sequence recombined into the NIb/CP junction which can accommodate heterologous genes in engineered infectious potyvirus clones. Homology of HAM1h with cellular Maf/HAM1 NTP pyrophosphatases suggests that HAMh1 might intercept non-canonical NTPs to reduce mutation rates of viral RNA.
Antigenic properties
Moderately immunogenic. No serological relationships with other members of the family Potyviridae have been found.
Biological properties
Host range
The natural host range of SPMMV is wide, with more than nine families susceptible, whereas the host range of CBSV, CVYV and SqVYV is less known apart from the hosts which they have been found to infect in the field.
Transmission
CBSV, CVYV and SqVYV are transmitted by the whitefly Bemisia tabaci in a non-persistent manner. B. tabaci may also be the vector of SPMMV, but this is not fully confirmed. All ipomoviruses are transmissible experimentally by mechanical inoculation and by grafting.
List of species in the genus Ipomovirus
| Cassava brown streak virus |
|
|
| Cassava brown streak virus-Tanzania:KOR6 | [GU563327] | (CBSV-KOR6) |
| Cucumber vein yellowing virus |
|
|
| Cucumber vein yellowing virus-Spain:ALM32 | [AY578085=NC_006941] | (CVYV-ALM32) |
| Squash vein yellowing virus |
|
|
| Squash vein yellowing virus-Florida | [EU259611=NC_010521] | (SqVYV-FL) |
| Sweet potato mild mottle virus |
|
|
| Sweet potato mild mottle virus-East Africa | [Z73124=NC_003797] | (SPMMV-EA) |
Species names are in italic script; names of strains, isolates and synonyms are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Ipomovirus but have not been approved as species
| Ugandan cassava brown streak virus | [FJ039520], [FN434109]* | (UCBSV) |
* Two strains sharing 87% nucleotide sequence identity.
Genus Macluravirus
Type species Maclura mosaic virus
Distinguishing features
Macluraviruses resemble members of the genus Potyvirus in their transmission by aphids but virions are slightly shorter. They form a distinct group in phylogenetic analyses and have different polyprotein consensus cleavage sites.
Virion properties
Morphology
Virions are flexuous filaments mostly 650–675 nm×13–16 nm.
Physicochemical and physical properties
Virion S20,w is 155–158S; density in CsCl is 1.31–1.33 g cm−3.
Nucleic acid
Virions contain one molecule of linear positive sense, ssRNA. RNA is about 8.0 kb.
Proteins
Macluraviruses have a single CP species of 33–34 kDa.
Genome organization and replication
Complete genomes of macluraviruses are not yet available. The aa sequences of macluravirus CPs show limited (14–23%) identity with CP sequences of some aphid-transmitted potyviruses. Macluraviruses show significant aa sequence identity in portions of the replicase protein with viruses in other genera of the family Potyviridae. The macluraviruses seem to have a genome organization and replication strategy typical of viruses in the family Potyviridae.
Antigenic properties
Moderately immunogenic. No serological relationships to members of the genus Potyvirus have been found except for a weak reaction between Maclura mosaic virus (MacMV) and bean yellow mosaic virus.
Biological properties
Host range
Current information suggests that most viruses have a narrow host range, infecting species in up to nine host families.
Transmission
The viruses are transmitted by aphids in a non-persistent manner and experimentally by mechanical inoculation.
List of species in the genus Macluravirus
| Alpinia mosaic virus |
|
|
| Alpinia mosaic virus-Taiwan | [AF499025*] | (AlpMV-TW) |
| Cardamom mosaic virus |
|
|
| Cardamom mosaic virus-India:Yelsur | [AF189125*] | (CdMV-YE) |
| Maclura mosaic virus |
|
|
| Maclura mosaic virus-UK | [U58771*] | (MacMV-UK) |
| Chinese yam necrotic mosaic virus |
|
|
| Chinese yam necrotic mosaic virus-Japan | [AB044386*] | (ChYNMV-JP) |
| Narcissus latent virus |
|
|
| Narcissus latent virus-New Zealand | [DQ450199*] | (NLV-NZ) |
| Ranunculus latent virus |
|
|
| Ranunculus latent virus-RN128 | [DQ152193*] | (RanLV-RN128) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Partial sequence including the coat protein; complete genome sequence not available.
List of other related viruses which may be members of the genus Macluravirus but have not been approved as species
| Yam chlorotic necrotic mosaic virus | [FM997910*] | (YCNMV) |
* Partial sequence including the coat protein; complete genome sequence not available.
Genus Rymovirus
Type species Ryegrass mosaic virus
Distinguishing features
This genus contains only three viruses presumably transmitted by host-adapted populations of eriophyid mite species in a semi-persistent manner. The rymoviruses share a reciprocal monophyletic relationship with species of the genus Potyvirus (see Figure 6).
Virion properties
Morphology
Virions are flexuous filaments 690–720 nm×11–15 nm in size.
Physicochemical and physical properties
Virion density in CsCl is 1.325 g cm-3 (for ryegrass mosaic virus, RGMV). Virion S20,w is 165–166S for most members.
Nucleic acid
Virions contain a single molecule of linear positive sense ssRNA with a 3′-poly(A) terminus. Virion RNA is about 9.5 kb in size.
Proteins
Rymoviruses have one type of CP, with a theoretical Mr of 35,482 Da and an apparent Mr estimated by Western blots of 45 kDa for RGMV.
Genome organization and replication
The complete genome sequences available for two isolates of RGMV and one each of Agropyron mosaic virus (AgMV) and Hordeum mosaic virus (HoMV) indicate that rymoviruses have a genome organization (see Figure 2) and replication strategy similar to other species of the Potyviridae with monopartite genomes.
Antigenic properties
Particles of most rymoviruses are moderately immunogenic. HoMV and AgMV are serologically related.
Biological properties
Host range
Most rymoviruses have limited but widespread host ranges within the family Gramineae but some have relatively narrow host ranges.
Transmission
Transmission by eriophyid mites and mechanical transmission have been reported for most members. The eriophyid mites transmitting rymoviruses are different from those transmitting tritimoviruses. The cereal rust mite Abacarus hystrix transmits both RGMV and AgMV, but only the former is efficiently transmitted. No vector is known for HoMV. Recent studies have revealed that host-associated populations of A. hystrix represent a species complex.
List of species in the genus Rymovirus
| Agropyron mosaic virus |
|
|
| Agropyron mosaic virus-USA:ND402 | [AY623626=NC_005903] | (AgMV-ND402) |
| Hordeum mosaic virus |
|
|
| Hordeum mosaic virus-ATCC PV81 | [AY623627=NC_005904] | (HoMV-PV81) |
| Ryegrass mosaic virus |
|
|
| Ryegrass mosaic virus-Denmark | [Y09854=NC_001814] | (RGMV-DK) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Rymovirus but have not been approved as species
None reported.
Genus Tritimovirus
Type species Wheat streak mosaic virus
Distinguishing features
Tritimoviruses are transmitted by mites (but different from those that transmit rymoviruses). They form a distinctive phylogenetic cluster.
Virion properties
Morphology
Virions are flexuous filaments 690–700 nm long.
Physicochemical and physical properties
Virion S20,w is 166S for wheat streak mosaic virus (WSMV).
Nucleic acid
Virions contain a positive sense ssRNA, about 9.4–9.6 kb in size, with a 3′-poly(A) terminus.
Proteins
The viral CP is a single peptide of about 349 aa for WSMV and 320 aa for brome streak mosaic virus (BrSMV). The Mr estimated by electrophoresis is about 42 kDa.
Genome organization and replication
The WSMV genome consists of 9384 nt excluding the 3′-terminal poly(A) tail. Sequence analysis reveals an ORF of 3035 aa. The structure and organization of the WSMV genome is similar to those of other members of the family Potyviridae (see Figure 2) except the bymoviruses. Most known potyvirus motifs are present in the polyprotein of WSMV. However, motifs in the putative helper-component and CP of BrSMV are incomplete or missing, which may account for different vector relations of the tritimoviruses. The WSMV CP sequence shows limited (22-25%) identity with CP sequences of some aphid-transmitted potyviruses. WSMV shows significant aa sequence identity with aphid-transmitted potyviruses in the cylindrical inclusion protein and portions of the nuclear inclusion proteins. WSMV RNA has been translated in vitro into several large proteins immunoprecipitable with WSMV CP antiserum, suggesting that WSMV uses a proteolytic processing strategy to express functional proteins such as the CP. Antiserum to TEV 58 kDa nuclear inclusion protein also reacts with in vitro translation products of WSMV. An in vitro translation product is precipitated with antiserum to HC-Pro helper component of an isolate of the species Tobacco vein mottling virus. Comparative sequence analyses show similarities with other members of the family Potyviridae, but these are limited to the nine mature proteins. WSMV is especially susceptible to proteinases in planta and has CP molecules of 42, 36 and 32 kDa; the two smaller proteins are parts of the 42 kDa protein.
Antigenic properties
Moderately immunogenic. WSMV and oat necrotic mottle virus (ONMV) are serologically related to each other, but not to the other members of the family Potyviridae.
Biological properties
Host range
The viruses only affect hosts in the Gramineae but while WSMV has a wide host range BrSMV and ONMV have narrow ones.
Transmission
WSMV and BrSMV are transmitted by eriophyid mites in a semi-persistent manner. HC-Pro of WSMV is required for mite transmission. All definitive tritimoviruses are transmissible experimentally by mechanical inoculation.
List of species in the genus Tritimovirus
| Brome streak mosaic virus |
|
|
| Brome streak mosaic virus-France-11-Cal | [Z48506=NC_003501] | (BrSMV-FR) |
| Oat necrotic mottle virus |
|
|
| Oat necrotic mottle virus-Type-NE | [AY377938=NC_005136] | (ONMV-NE) |
| Wheat Eqlid mosaic virus |
|
|
| Wheat Eqlid mosaic virus-Iran | [EF608612=NC_009805] | (WEqMV-IR) |
| Wheat streak mosaic virus |
|
|
| Wheat streak mosaic virus-Sidney 81 | [AF057533=NC_001886] | (WSMV-Sidney 81) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Tritimovirus but have not been approved as species
| Yellow oat-grass mosaic virus | [GQ259764*] | (YOgMV) |
* Partial sequence including the coat protein; complete genome sequence not available.
Genus Bymovirus
Type species Barley yellow mosaic virus
Distinguishing features
Compared with other viruses in the family, members of the genus Bymovirus are distinct in having a divided (bipartite) genome and in being transmitted by the root-infecting parasite, Polymyxa graminis (Plasmodiophorales, a fungoid protist).
Virion properties
Morphology
Virions are flexuous filaments of two modal lengths, 250–300 nm and 500-600 nm; both are 13 nm in width (Figure 4).
Physicochemical and physical properties
Virion density in CsCl is 1.28–1.30 g cm−3.
Nucleic acid
Virions contain two molecules of linear positive sense, ssRNA. RNA-1 is 7.5–8.0 kb and RNA-2 is 3.5–4.0 kb; RNA makes up 5% by weight of particles. There is little base sequence identity between the two RNAs except in the 5′ UTR.
Proteins
Virions have a single CP of 28.5–33 kDa. The CP of barley yellow mosaic virus isolates has 297 aa.
Genome organization and replication
The two RNA molecules are translated initially into precursor polypeptides from which functional proteins are derived by proteolytic processing (Figure 5). The organization of RNA1 is similar to that of other potyviruses but without the P1 and HC-Pro proteins. The RNA2 polyprotein is unique to bymoviruses although the first (ca. 28 kDa) protein from RNA-2 has aa domains with sequence similarities to the potyvirus protein HC-Pro. The larger protein of RNA2 is believed to have a role in vector transmission.
Antigenic properties
The viral proteins are moderately immunogenic; serological relationships exist among most members (except barley mild mosaic virus). The CP aa sequence identity among members is 35–74%.
Biological properties
Cytology
There are characteristic pinwheel-like inclusions and membranous network structures are formed in the cytoplasm of infected plant cells. No nuclear inclusions are found.
Host range
The host range of member viruses is narrow, restricted to the host family Gramineae. Each species has a very restricted host range; for example, the barley-infecting species do not infect wheat and vice versa.
Transmission
These viruses are transmitted by Polymyxa graminis in a persistent manner, surviving in resting spores as long as these remain viable; it is transmissible experimentally by mechanical inoculation, sometimes with difficulty.
List of species in the genus Bymovirus
| Barley mild mosaic virus |
|
|
| Barley mild mosaic virus-fungally transmissible UK isolate | [Y10973=NC_003483, X90904=NC_003482,] | (BaMMV-F) |
| Barley yellow mosaic virus |
|
|
| Barley yellow mosaic virus-IIa | [D01091-2] | (BaYMV-IIa) |
| Oat mosaic virus |
|
|
| Oat mosaic virus-UK | [AJ306718-9=NC_004016-7] | (OMV-UK) |
| Rice necrosis mosaic virus |
|
|
| Rice necrosis mosaic virus-Japan | [U95205*] | (RNMV-JA) |
| Wheat spindle streak mosaic virus |
|
|
| Wheat spindle streak mosaic virus-France | [X73883*] | (WSSMV-FR) |
| Wheat yellow mosaic virus |
|
|
| Wheat yellow mosaic virus-Japan | [D86634-5=NC_002349-50] | (WYMV-JA) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Partial sequence of RNA1, including the coat protein; complete genome sequence not available.
List of other related viruses which may be members of the genus Bymovirus but have not been approved as species
None reported.
List of unassigned species in the family Potyviridae
| Spartina mottle virus |
|
|
| Spartina mottle virus-Germany | [AF491351*] | (SpMoV-DE) |
| Sugarcane streak mosaic virus |
|
|
| Sugarcane streak mosaic virus-Pakistan | [GQ388116] | (SCSMV-PAK) |
| Tomato mild mottle virus |
|
|
| Tomato mild mottle virus-Yemen | [AF359575*] | (TomMMoV-YM) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Partial sequence including the coat protein; complete genome sequence not available.
Tomato mild mottle virus is most closely related to members of the genus Ipomovirus but is transmitted by aphids in a non-circulative manner, whereas ipomoviruses are transmitted by the whitefly Bemisia tabaci. No vectors have been identified for the other unassigned members of the family.
List of other related viruses which may be members of the family Potyviridae but have not been approved as species
| Triticum mosaic virus-Kansas isolate U06-123 | [FJ263671=NC_012799] | (TriMV) |
There is a current proposal to make Triticum mosaic virus, recently described in the USA and transmitted by the same mite vector as tritimoviruses, the type species of a new genus, Poacevirus. If accepted, this genus would also contain the unassigned species Sugarcane streak mosaic virus, which is clearly related.
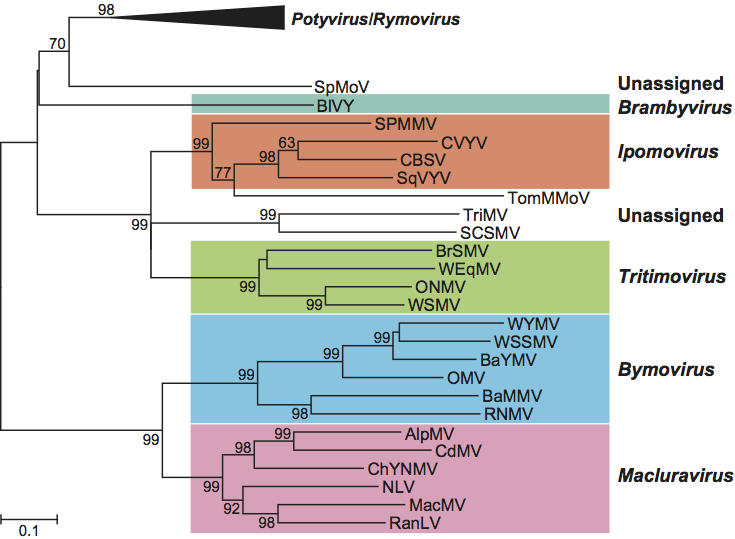
Phylogenetic relationships within the family
Phylogenetic relationships within the family are depicted in Figures 6 and 7.
Similarity with other taxa
Viruses in the family Potyviridae have similarity to members of the order Picornavirales. In particular, the genomes have a VPg at their 5′ termini and a poly(A) tract at their 3′ termini. Their genomes are expressed initially as high molecular weight polyprotein precursors which are processed by virus-encoded proteases. Gene products involved in replication are conserved in gene order and gene sequence. However, members of the Picornavirales have isometric particles, a much smaller VPg and a different type of helicase. There are also some sequence similarities to members of the family Hypoviridae.
Derivation of names
Bramby: from bramble, the host of the type species.
Bymo: from barley yellow mosaic.
Ipomo: from Ipomea and mosaic.
Maclura: the genus name of the host of the type species.
Poty: from potato virus Y.
Rymo: from ryegrass mosaic.
Tritimo: from Triticum and mosaic.
Further reading
Adams et al., 2005 M.J. Adams, J.F. Antoniw, F. Beaudoin, Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol. Plant Pathol. 6 (2005) 471–487.
Adams et al., 2005 M.J. Adams, J.F. Antoniw, C.M. Fauquet, Molecular criteria for genus and species discrimination within the family Potyviridae. Arch. Virol. 150 (2005) 459–479.
Chung et al., 2008 B.Y.W. Chung, W.A. Miller, J.F. Atkins, E.A. Firth, An overlapping essential gene in the Potyviridae. Proc. Natl Acad. Sci., U S A. 105 (2008) 5897–5902.
French and Stenger, 2005 R. French, D.C. Stenger, Genome sequences of Agropyron mosaic virus and Hordeum mosaic virus support reciprocal monophyly of the genera Potyvirus and Rymovirus in the family. Potyviridae. Arch. Virol. 150 (2005) 299–312.
Gibbs and Ohshima, 2010 A. Gibbs, K. Ohshima, Potyviruses and digital revolution. Annu. Rev. Phytopathol. 48 (2010) 10.1–10.19.
Kekarainen et al., 2002 T. Kekarainen, H. Savilahti, J.P.T. Valkonen, Functional genomics on Potato virus A: a virus genome-wide map of sites essential for virus propagation. Genome Res. 12 (2002) 584–594.
Kelloniemi et al., 2008 J. Kelloniemi, K. Mäkinen, J.P.T. Valkonen, Three heterologous proteins simultaneously expressed from a chimeric potyvirus: infectivity, stability and the correlation of genome and virion lengths. Virus Res. 135 (2008) 282–291.
Rajamäki et al., 2004 M.L. Rajamäki, T. Mäki-Valkama, K. Mäkinen, J.P.T. Valkonen, N.J. Talbot, Infection with potyvirusesPlant–Pathogen Interactions. In: N.J. Talbot, Plant–Pathogen Interactions. Blackwell Publishing, Sheffield, UK200468–91.
Susaimuthu et al., 2008 J. Susaimuthu, I.E. Tzanetakis, R.C. Gergerich, R.R. Martin, A member of a new genus in the Potyviridae infects Rubus. Virus Res. 131 (2008) 145–151.
Valli et al., 2007 A. Valli, J.J. López-Moya, J.A. García, Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae. J. Gen. Virol. 88 (2007) 1016–1028.
Winter et al., 2010 S. Winter, M. Koerbler, B. Stein, A. Pietruszka, M. Paape, A. Butgereitt, Analysis of cassava brown streak viruses reveals the presence of distinct virus species causing cassava brown streak disease in East Africa. J. Gen. Virol. 91 (2010) 1365–1372.
Contributed by
Adams, M.J., Zerbini, F.M., French, R., Rabenstein, F., Stenger, D.C. and Valkonen, J.P.T.
Figures
Figure 1 (Left) Schematic diagram of a potyvirus particle. The N-terminal (ca. 30 aa; large rectangle) and C-terminal (ca.19 aa; small rectangle) of the CP molecules are exposed on the surface of the intact virus particle (from Shukla and Ward (1989). Adv. Virus Res., 36, 273-314). (Right) Negative contrast electron micrograph of particles of an isolate of Plum pox virus, stained with 1% PTA, pH 6.0. The bar represents 200 nm.
(Courtesy of I.M. Roberts.)
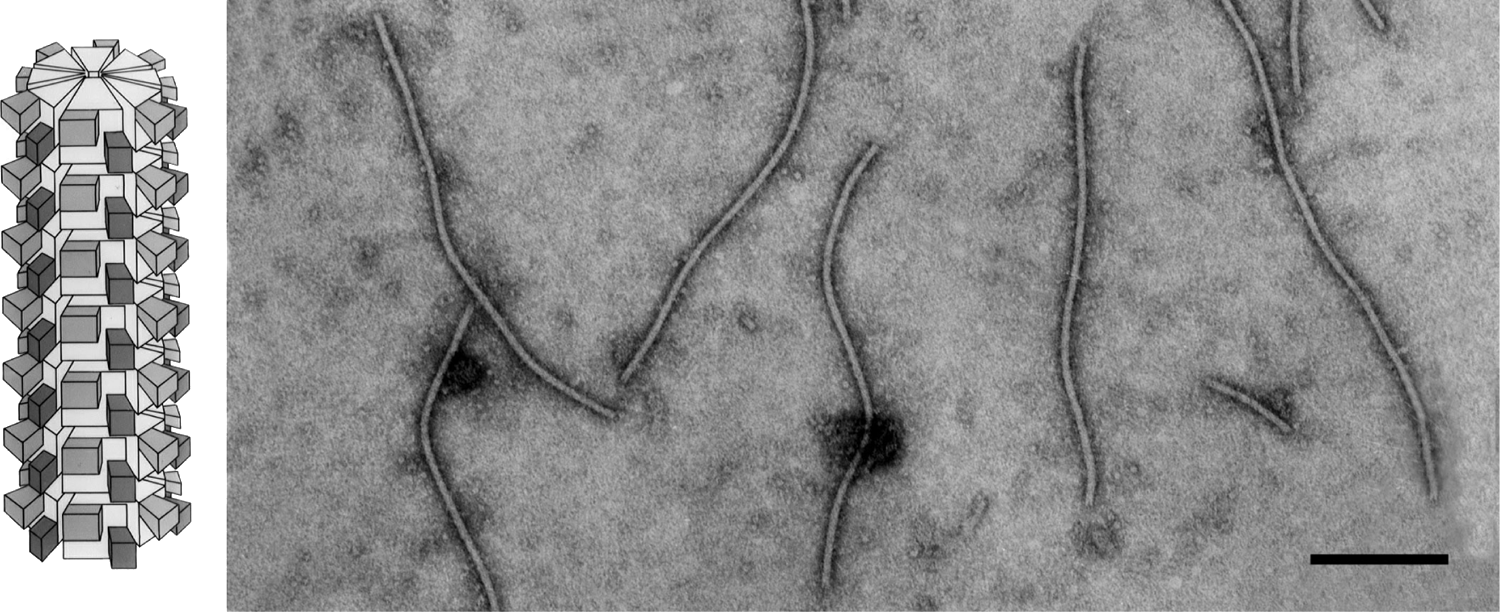
Figure 2 Genomic map of a member of the genus Potyvirus, using a strain of Tobacco etch virus as an example. The ssRNA genome is represented by a line and an open box representing the ORF translated into a polyprotein. Functions associated with the mature proteins proteolytically processed from the polyprotein are shown. VPg, genome-linked viral protein covalently attached to the 5-terminal nt (represented by the oval at the 5 end); P1-Pro, a protein with a proteolytic activity responsible for cleavage at typically Tyr/Phe-Ser (); HC-Pro, a protein with aphid transmission helper-component activity and proteolytic activity responsible for cleavage at typically Gly-Gly (); Pro, serine-like proteolytic activity responsible for cleavage at Gln/Glu-(Ser/Gly/Ala) (). Some of these proteins of particular viruses of the family Potyviridae aggregate to form inclusion bodies during infection. The protein involved and the particular type of inclusion body is shown above the genetic map; AI, amorphous inclusion; CI, cylindrical-shaped inclusion body found in the cytoplasm; NIa and NIb, small and large nuclear inclusion proteins, respectively, which aggregate in the nucleus to form a nuclear inclusion body. The small ORF PIPO is putatively translated by +2 frameshift of the polyprotein ORF, and its product is expressed as a fusion with the N-terminal part of P3.
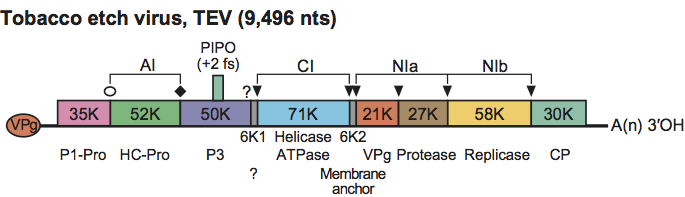
Figure 3 Genomic maps of the ipomoviruses sweet potato mild mottle virus (SPMMV), squash vein yellowing virus (SqVYV) and cassava brown streak virus (CBSV). The ssRNA genome is represented by a line and an open box representing the ORF translated into a polyprotein. Conventions are as for the potyvirus genome organization map (Figure 2). Activities of most gene products are postulated by analogy with genus Potyvirus. CVYV and SqVYV contain two P1-like serine proteases (P1a and P1b), of which P1b functions as a suppressor of RNA silencing. CBSV also contain P1b which suppresses silencing and, additionally, carries a Maf/HAM1-like sequence recombined into the NIb/CP junction. HAMh1 might intercept non-canonical NTPs to reduce mutation rates of viral RNA.
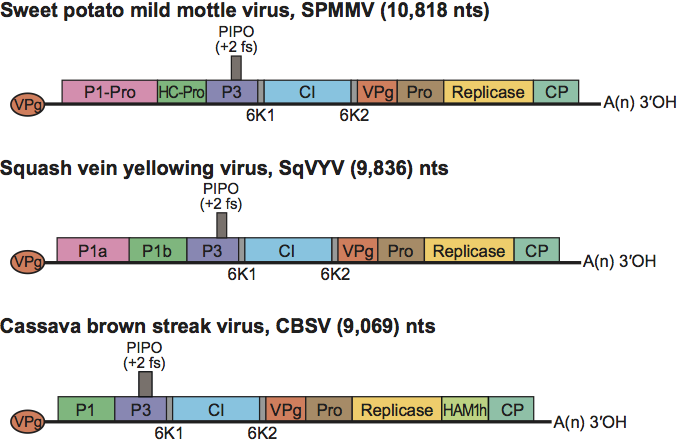
Figure 4 Virions of an isolate of barley yellow mosaic virus, stained with 1% PTA, pH 7.0. The bar represents 200 nm (from D. Lesemann).
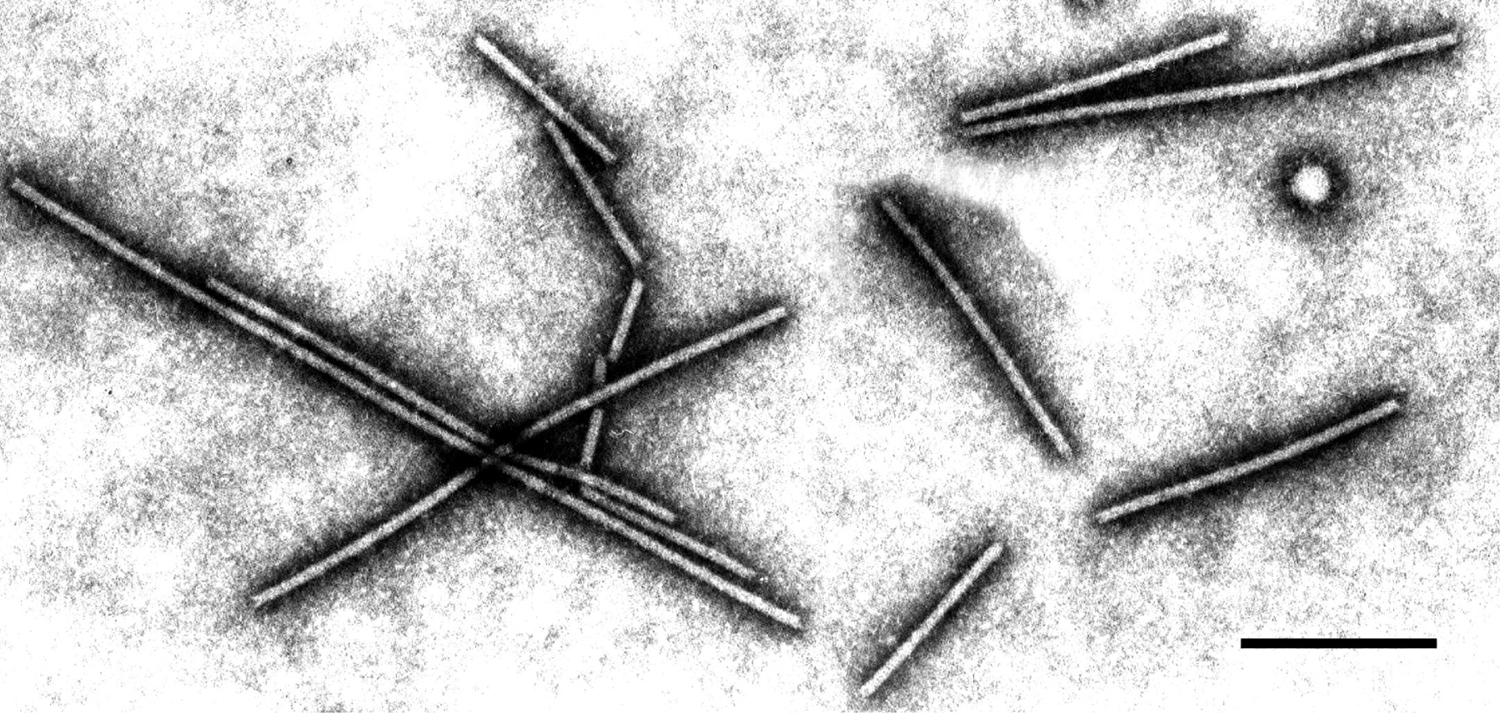
Figure 5 Genomic map of the bymovirus bipartite genome, using, as an example an isolate of barley yellow mosaic virus. Conventions are as for potyvirus genome organization map (Figure 2). Function of most gene products are postulated by analogy with genus Potyvirus. P1 corresponds to the C-terminal protease of HC-Pro.
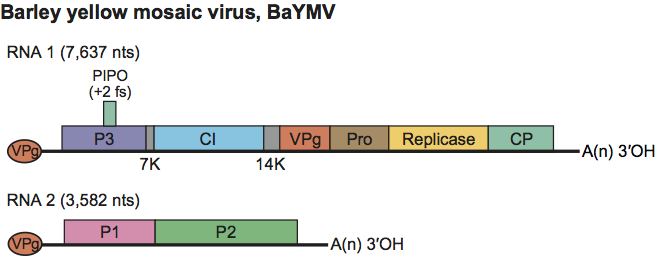
Figure 6 Unrooted phylogenetic tree based on the codon-aligned nucleotide sequences of the polyproteins of fully-sequenced members of the family Potyviridae. The tree uses the majority of the polyprotein (from the 6K1 cistron to the end of the coat protein). One representative sequence was chosen for each species. The analysis was done in MEGA4 (maximum composite likelihood distances) and the numbers on major branches indicate percentage of bootstrap support out of 10,000 bootstrap replications.
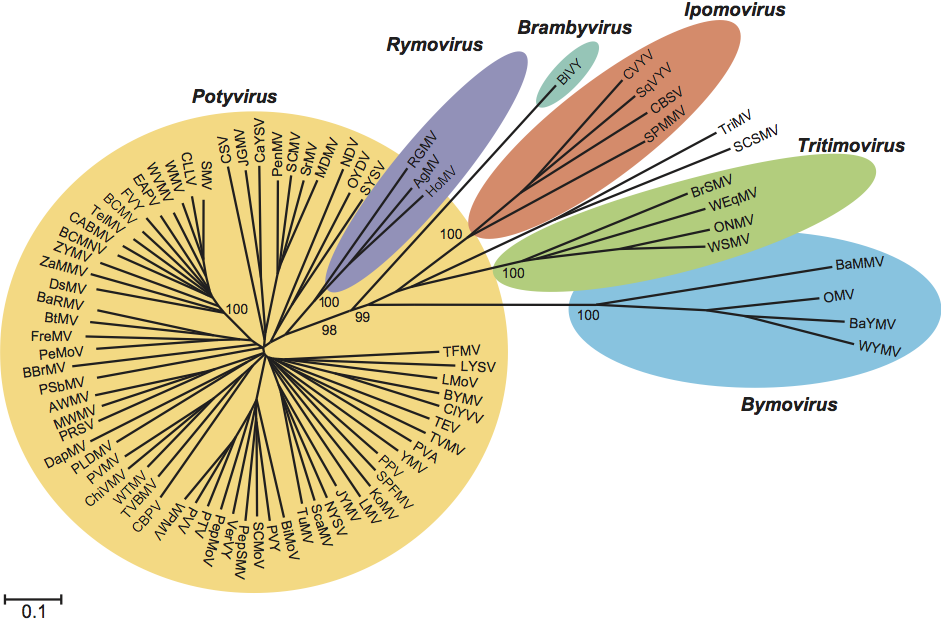
Figure 7 Phylogenetic (distance) tree based on the codon-aligned nucleotide sequences of the 3 ends (partial NIb and complete coat protein) of the polyproteins of members of the family Potyviridae. The analysis was done in MEGA4 (maximum composite likelihood distances) and the numbers on branches indicate percentage of bootstrap support out of 10,000 bootstrap replications (where >60%). Because of the large number of sequences in the genus Potyvirus, the branch for this genus (and Rymovirus) has been collapsed. The tree is provided particularly to demonstrate the position of unassigned members of the family and those in the genus Macluravirus, where complete sequences are not available.