Genus: Polemovirus
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Type species: Poinsettia latent virus
Distinguishing features
This is a monotypic genus containing a virus that has a close relationship to poleroviruses within the first three-quarters of its genome but to sobemoviruses in the last quarter.
Virion properties
Morphology
Virions are 34 nm in diameter, hexagonal in outline and have no envelope. They are similar to those of sobemoviruses and exhibit icosahedral symmetry (T=3).
Physicochemical and physical properties
These have not been studied but are probably similar to those of sobemoviruses.
Nucleic acid
Particles contain a single molecule of positive sense ssRNA, about 4.6 kb in size. A 1.07 kb subgenomic RNA (sgRNA) molecule, co-terminal with the 3′ end of the genomic RNA, can be detected in infected cells and a larger sgRNA of 2.60 kb may also be present. The 3′ terminus is non-polyadenylated and is predicted to form a stable hairpin. It is likely that a VPg protein is covalently linked to the 5′ end of the genomic RNA.
Proteins
There is one major structural protein of 33 kDa. A genome-linked (VPg) protein is predicted from the sequence data and by comparison with poleroviruses, but has not been detected in vivo.
Lipids
None reported.
Carbohydrates
None reported.
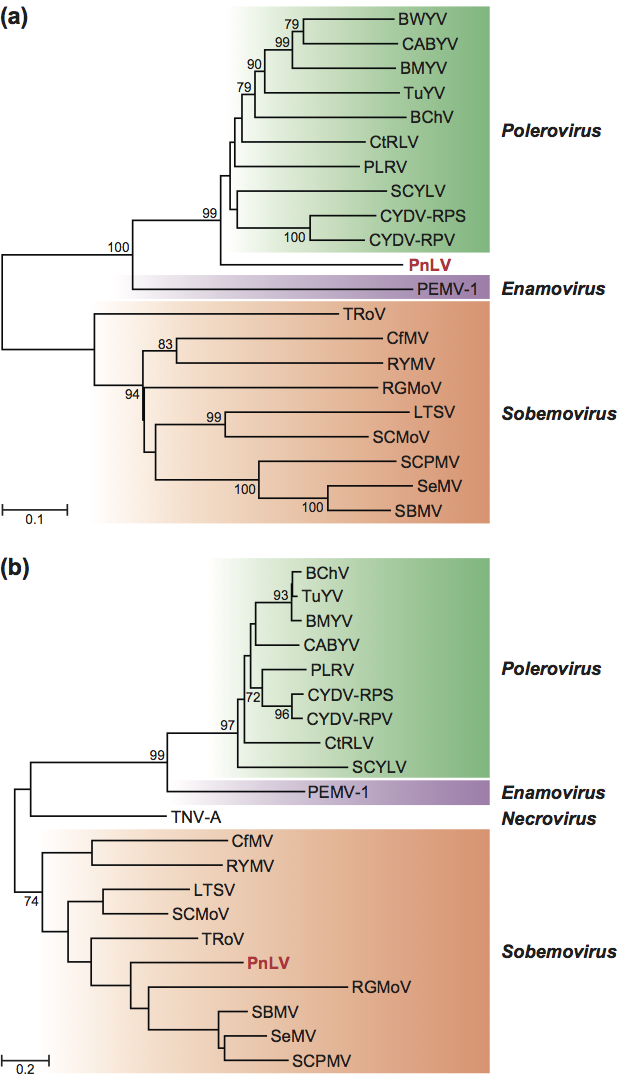
Genome organization and replication
Four ORFs have been identified in the sequence determined (Figure 1). The first three ORFs resemble those of members of the genus Polerovirus (family Luteoviridae) and have been labelled similarly. The ORF0 product is therefore probably a suppressor of gene silencing. ORF1 contains helicase, putative VPg and protease domains, while ORF2 is an RNA-dependent RNA polymerase. As in poleroviruses, it is likely that ORFs 0, 1 and 2 are translated from the genomic RNA and that ORF2 is translated by frameshift from ORF1, thus sharing the same amino terminus. ORF3 encodes the viral coat protein and is believed to be translated from the sgRNA that can be detected in infected plants.
Antigenic properties
Virions are strongly immunogenic.
Biological properties
The virus occurs regularly in cultivated poinsettia plants throughout the world, apparently without causing symptoms. It is distributed by grafting and vegetative propagation of the host plant but its natural means of transmission is unknown. Its appearance in virus-free plants has lead to suggestions that transmission from soil may occur. It often occurs in association with poinsettia mosaic virus but the two viruses are serologically and genetically unrelated.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Polemovirus
| Poinsettia latent virus |
|
|
| Poinsettia latent virus |
|
|
| Poinsettia latent virus-Germany | [AJ867490=NC_011543] | (PnLV-DE) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Polemovirus but have not been approved as species
None reported.
Similarity with other taxa
The virus was initially named poinsettia cryptic virus but has now been shown to be unrelated to the cryptic viruses, which have dsRNA genomes. Sequence comparisons show that the 5′-section of the genome is similar in organization and sequence to members of the genus Polerovirus (family Luteoviridae), whereas the coat protein is most closely related to members of the genus Sobemovirus (Figure 2). The virus therefore probably arose by recombination although the exact history may be more involved. There is a complex network of putative recombination amongst members of the families Luteoviridae and Tombusviridae and the unassigned genera Polemovirus and Sobemovirus that is difficult to reflect by conventional hierarchical taxonomy.
Derivation of name
Polemo: from genera Polerovirus and Sobemovirus to indicate the chimeric nature of the virus.
Further reading
Koenig and Lesemann, 1980 R. Koenig, D.E. Lesemann, Two isometric viruses in Poinsettias. Plant Dis. 64 (1980) 782–784.
Siepen et al., 2005 M. Siepen, S. aus dem, J.O. Pohl, B.J. Koo, C. Wege, H. Jeske, Poinsettia latent virus is not a cryptic virus, but a natural polerovirus–sobemovirus hybrid. Virology. 336 (2005) 240–250.
Contributed by
Adams, M.J. and Jeske, H.
Figures
Figure 1 Diagram of the genome organization and map of the translation products of poinsettia latent virus.
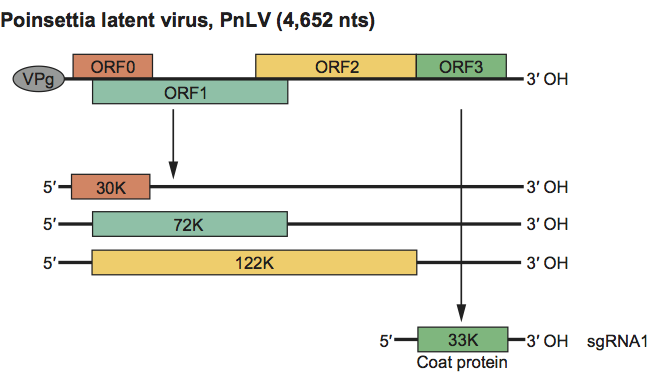
Figure 2 Phylogenetic (distance) trees of (a) the RdRp domain and (b) the coat protein nucleotide sequences of members of the genera Polerovirus and Sobemovirus showing the different placement of poinsettia latent virus (PnLV) in the two regions of the genome. Trees were prepared from codon-aligned sequences in MEGA4 (maximum composite likelihood distances and 10,000 bootstrap replicates). Bootstrap values >70% are shown at the nodes.