Genus: Ourmiavirus
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Type species: Ourmia melon virus
Distinguishing features
Plant viruses with unenveloped bacilliform virions composed of a single coat protein and three positive sense ssRNAs. Only three proteins are expressed. The virions possess a unique fine structure with a series of discrete lengths from 30 to 62 nm. The RdRp has closest similarity to that of the fungal virus genus Narnavirus (Narnaviridae); the movement protein is significantly similar to the MPs of tombusviruses (Tombusviridae); the coat protein shows limited similarity to the CPs of several plant and animal viruses. This combination of characters is not found in any other virus taxon.
Virion properties
Morphology
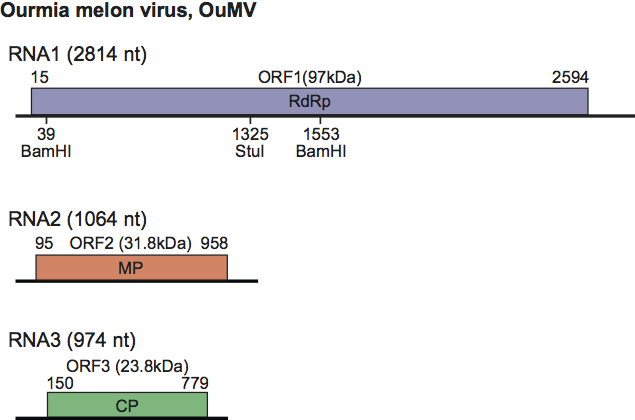
The bacilliform virions constitute a series of particles with conical ends (apparently hemi-icosahedra) and cylindrical bodies 18 nm in diameter. The bodies of the particles are composed of a series of double disks, the commonest particle having two disks (particle length 30 nm), a second common particle having three disks (particle length 37 nm) with rarer particles having four disks (particle length 45.5 nm) and six disks (particle length 62 nm). There is no envelope. (See Figures 1 and 2.)
Physicochemical and physical properties
The Mr of virions and their sedimentation coefficients are not known. The buoyant density in CsCl of all particle sizes is 1.375 g cm−3. The particles are stable near pH 7. Thermally, they are relatively stable; infectivity survives in crude sap after heating for 10 min at 70 °C but not 80 °C, and is retained after at least one freeze–thaw cycle. The particles survive in CsCl density gradients and survive treatment with chloroform but not n-butanol, and survive treatment with 1% Triton X-100 detergent.
Nucleic acid
There are three positive sense ssRNAs. In Ourmia melon virus (OuMV) these have sizes of 2814, 1064 and 974 nt, with similar values for the other members of the genus.
Proteins
For OuMV there is one structural protein of 23.8 kDa; the other two proteins are the RdRp (97.5 kDa) and the movement protein (31.6 kDa). Data are similar for the other two species.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Each RNA has one ORF (Figure 3). RNA-1 encodes a protein carrying the GDD motif typical of RNA-dependent RNA polymerases (RdRps). There is evidence that RNA-2 encodes the cell-to-cell movement protein. The product of RNA-3 is the coat protein, which localizes to the nucleus. Synthesis of coat protein from actively replicating RNA-3 is necessary for both virion assembly and systemic infection of the host. There appear to be no subgenomic RNAs and no further proteins produced by read through. The movement protein may undergo post-translational modification. Details of replication are not known except that virions and the movement protein accumulate in the cytoplasm.
Antigenic properties
The virions (i.e. the assembled coat protein) are good immunogens, as are the tubular structures associated with the movement protein. Antisera to these proteins did not react in Western blots to extracts of plants infected with either of the other two viruses in the genus.
Biological properties
The type species can easily be mechanically transmitted to a rather wide range of dicot plants (about 40 species in 15 families reported), usually inducing systemic ringspots, mosaic and necrosis, with local lesions on some hosts. There is no particular tissue tropism. No vector has been identified but several species of weeds around infected fields are commonly also infected. No experimental transmission was obtained with several aphid species, the whiteflies Trialeurodes and Bemisia or a Tetranychus mite. Attempts at transmission through soil or irrigation water were negative. Experimental seed-transmission rates are 1–2% in Nicotiana benthamiana and N. megalosiphon. Different species in the genus occur in geographically diverse areas and on widely different hosts, though there are experimental hosts in common.
Species demarcation criteria in the genus
- Not related serologically with respect to both the coat protein and the movement protein.
- Natural hosts are widely different.
- Less than 60% similarity between species in the coat protein aa sequence.
List of species in the genus Ourmiavirus
| Ourmia melon virus |
|
|
| Ourmia melon virus-VE9 | [RNA-1: EU770623; RNA-2: EU770624; RNA-3: EU770625] | (OuMV-VE9) |
| Epirus cherry virus |
|
|
| Epirus cherry virus-VE450 | [RNA-1: EU770620; RNA-2: EU770621; RNA-3: EU770622] | (EpCV-VE450) |
| Cassava virus C |
|
|
| Cassava virus C-IC | [RNA-1: FJ157981; RNA-2: FJ157982; RNA-3: FJ157983] | (CsVC-IC) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Ourmiavirus but have not been approved as species
None reported.
Similarity with other taxa
The genus Ourmiavirus stands isolated, having particles of unique morphology and a unique combination of phylogenetic affinities for the three RNAs. The RdRp encoded by RNA-1 has closest (although distant) affinity with the RdRp of the fungal virus Narnavirus (Narnaviridae). The movement protein encoded by RNA-2 has clear affinities with the MPs of viruses in the Tombusviridae. The coat protein shows distant affinities with the CPs of sobemo-, tombus- and luteoviruses (plant viruses) and nodaviruses (animal viruses).
Derivation of names
Ourmia: from Ourmia (Urmia, Orumieh) in north-western Iran where the type virus was first found.
Epirus: from Epirus, Greece, where cherry virus was isolated.
Cassava: cassava virus C has been isolated from the cassava plant in various parts of Africa.
Further reading
Aiton, M.M., Roberts, I.M. and Harrison, B.D. (1988). Two new cassava viruses from Africa. Abstracts 5th Int. Congress Plant Pathol., Kyoto, 1988, p. 43.
Avgelis, A., Barba, M. and Rumbos, I. (1989). Epirus cherry virus, an unusual virus isolated from cherry with rasp-leaf symptoms in Greece. J. Phytopathol., 126, 51-58.
Crivelli, G., Ciuffo, M., Genre, A., Masenga, V. and Turina, M. (2011). Reverse genetic analysis of ourmiaviruses reveals the nucleolar localization of the coat protein in Nicotiana benthamiana and unusual requirements for virion formation. J. Virol., 85, in press.
Lisa, V., Milne, R.G., Accotto, G.P., Boccardo, G., Caciagli, P. and Parvizy, R. (1988). Ourmia melon virus, a virus from Iran with novel properties. Ann. Appl. Biol., 112, 291-302.
Rastgou, M., Habibi, M.K., Izadpanah, K., Masenga, V., Milne, R.G., Wolf, Y.I., Koonin, E.V. and Turina, M. (2009). Molecular characterization of the plant virus genus Ourmiavirus and evidence of inter-kingdom reassortment of viral genome segments as its possible route of origin. J. Gen. Virol., 90, 2525-2536.
Contributed by
Rastgou, M., Turina, M. and Milne, R.G.
Figures
Figure 1 Diagram of virion surface of a member of the genus Ourmiavirus, showing arrangement of double disks and conical ends in particles of different length. Each row of five triangles represents a double disk.

Figure 2 (A, B, C) Negative contrast electron micrographs (uranyl acetate) of purified particles of Ourmia melon virus. The bar represents 100 nm. D and E, features of the two commonest particle types, enhanced by photographic superimposition.
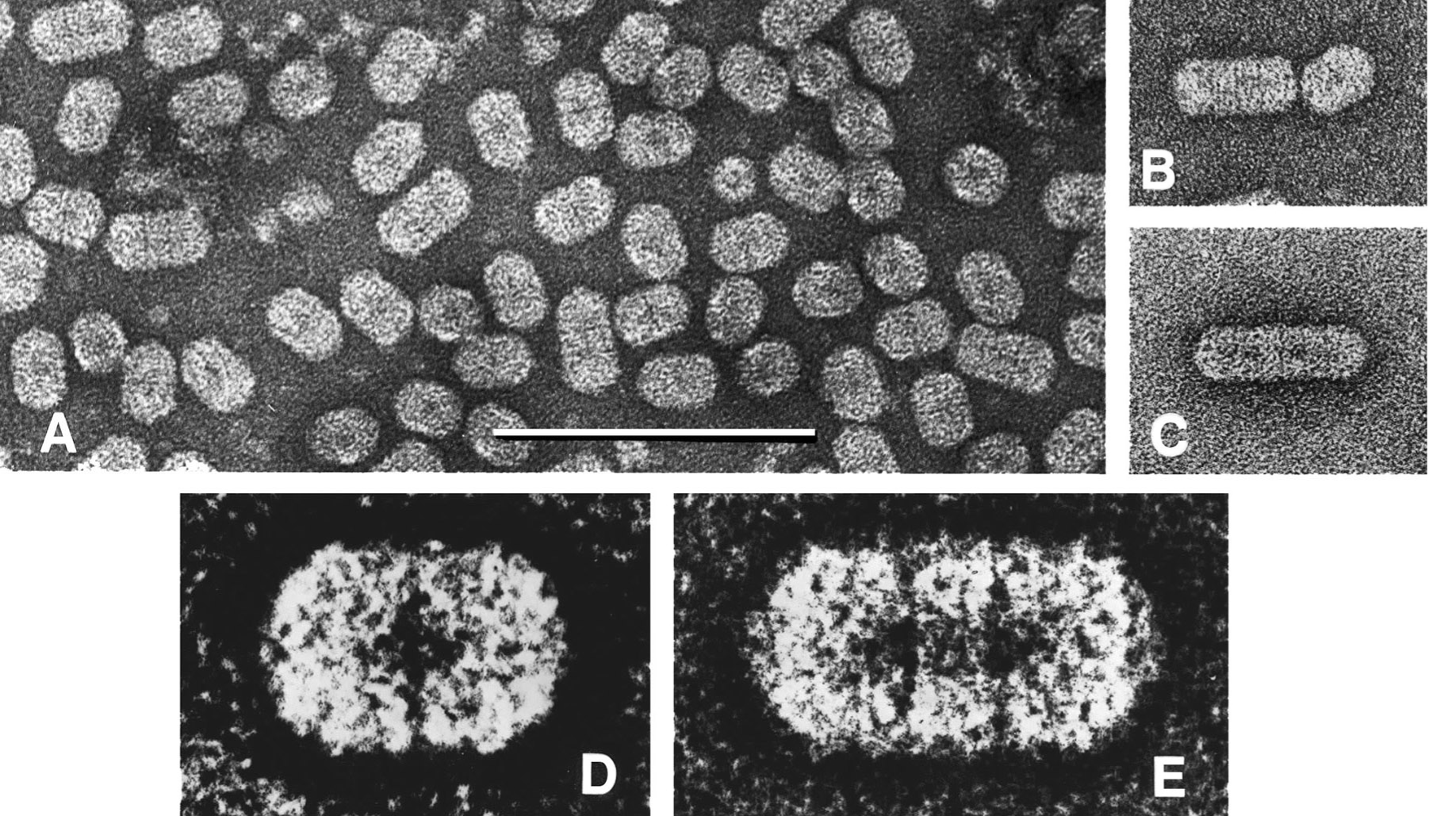
Figure 3 Diagram of the genome organization of Ourmia melon virus isolate VE9 showing the size of each RNA and the positions and sizes of the ORFs. CP, coat protein; MP, movement protein; RdRp, RNA-dependent RNA polymerase.