Family: Nodaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Genus Alphanodavirus
Type species Nodamura virus
Virion properties
Morphology
Virions are non-enveloped, roughly spherical in shape, 32–33 nm in diameter and have icosahedral symmetry (T=3). No distinct surface structure is seen by electron microscopy of negatively stained preparations. Empty shells are seldom seen in virus preparations. (See Figure 1.)
Physicochemical and physical properties
Virion Mr is about 9×106; S20,w is 135–145S. Virion buoyant density in CsCl is 1.30–1.34 g cm−3 (varies with species). Infectivity of aqueous suspensions is stable to extraction with chloroform. Infectivity of Nodamura virus (NoV), black beetle virus (BBV), or Flock House virus (FHV) is stable at room temperature in 1% sodium dodecyl sulfate but Boolarra virus (BoV) is inactivated. Virions are stable at acid pH. The RNA content of the virion is about 16%.
Nucleic acid
The genome consists of two molecules of positive sense ssRNA: RNA1 (Mr 1.1×106, 3.1 kb) and RNA2 (Mr 0.48×106, 1.4 kb). Both molecules are required for infectivity, and both are encapsidated in the same virus particle. Both RNA molecules are capped at their 5′ ends with cap zero structures and lack poly(A) tails at their 3′ ends. RNA 3′ ends cannot be chemically derivatized even after treatment with denaturing solvents, indicating that the expected 3′-terminal-OH groups are unreactive.
Proteins
The capsid consists of 180 protein subunits (protomers) arranged on a T=3 surface lattice. Each protomer is composed of a single CP (protein α) or the two products of its cleavage (proteins β and γ). Mass spectrometry of the FHV CP indicates that the initiating methionine is removed. Thus, for FHV, the capsid proteins are: protein α: (44 kDa), aa 2-407; protein β: (39 kDa), aa 2–363; protein γ: (4 kDa), aa 364–407. Morphogenesis involves the formation of a non-infectious provirion, which acquires infectivity by autocatalytic cleavage of protein α to form proteins β and γ. Maturation cleavage is often incomplete and virions typically contain residual uncleaved protein α.
Lipids
None.
Carbohydrates
None.
Genome organization and replication
Alphanodaviruses replicate in the cytoplasm of infected cells (Figure 2). RNA synthesis is resistant to actinomycin D. Infected cells contain three ssRNAs: RNA1 (Mr 1.1×106; 3.1 kb); RNA2 (Mr 0.48×106; 1.4 kb) and a sgRNA3 (Mr 1.10.13×106; 0.39 kb), whose nt sequence corresponds to the 3’ end of RNA1 (387 nt in the case of FHV). The 3’ end of all three RNAs is chemically unreactive. Unlike RNAs 1 and 2, RNA3 is not packaged into virions. RNA1 encodes protein A (112 kDa), which is the catalytic subunit of the viral RdRp. RNA2 encodes protein α, the CP precursor (44 kDa). Depending on the virus species, RNA3 encodes one or two small proteins (proteins B1 and B2, 11 kDa). B1 is encoded in the same ORF as protein A. Protein B2 is encoded in an overlapping ORF. BoV RNA3 does not encode protein B1 but all known alphanodavirus RNA3 molecules encode protein B2. Protein B2 of FHV functions as a suppressor of RNA interference (RNAi) in Drosphila melanogaster, cultured Drosophila melanogaster cells (Schneider’s line 2), tobacco plants (Nicotiana benthamiana), and Caenorhabditis elegans. Similarly, NoV B2 suppresses RNAi in cultured mammalian and mosquito cells. The function of protein B1 is unknown. Cells transfected with isolated RNA1 synthesize RNA1 and overproduce RNA3, but do not make RNA2. RNA2 replication strongly inhibits synthesis of RNA3 and the translation of RNA2 suppresses the translation of RNA1.
Antigenic properties
NoV, BBV, FHV and BoV are cross-reactive by double-diffusion immunoprecipitation tests, but all four members represent different serotypes (neutralization titer of each antiserum less than 0.5% in heterotypic crosses). In constrast, PaV and NoV are not cross-reactive by gel immunodiffusion tests.
Biological properties
Host range
Nature: All species of alphanodaviruses were isolated in nature from insects, although serological data suggest that NoV also naturally infects pigs and perhaps herons. NoV seems to be unique among the nodaviruses in its ability to infect both vertebrates and invertebrates. It is also very unusual in being able to kill both insect and mammalian hosts. The other alphanodaviruses do not show strict specificity for particular insect hosts.
Laboratory: In the laboratory, most alphanodaviruses can be propagated in larvae of the common wax moth, Galleria mellonella, where they cause paralysis and death. FHV, BBV, and BoV grow well in cultured Drosophila melanogaster cells and form plaques on monolayers of these cells. Defective-interfering particles are readily formed unless the viruses are passaged at low multiplicity of infection. FHV, isolated from grass grubs (Costelytra zealandica), also infects several other insect species, including adult common fruit flies (Drosophila melanogaster), tsetse flies (Glossina morsitans morsitans), reduviid bugs (Rhosnius prolixus) and several mosquito species (Aedes aegypti, Culex pipiens pipiens, Armigeres subalbatus and Anopheles gambiae). FHV can be propagated in mammalian cells, plants (Nicotiana benthamiana), yeast (Saccharomyces cerevisiae) and nematodes (Caenorhabditis elegans). Persistent FHV infections, with subsequent resistance to superinfection, occur readily in cultured Drosophila melanogaster cells. NoV, isolated from mosquitoes, also causes paralysis and death in suckling mice and suckling hamsters. NoV infects cultured cells from both mosquito and mammalian species, but not those of Drosophila melanogaster. Interestingly, NoV infection is delayed in mammalian cells compared to that seen in mosquito cells. NoV can also be propagated by transfecting mosquito, vertebrate, or yeast (Saccharomyces cerevisiae) cell cultures with virion RNA or cloned cDNA copies of genomic RNAs at temperatures up to 37 °C. PaV infects cultured cells from Spodoptera exigua (beet armyworm), Helicoverpa zea (corn earworm), and Aedes albopictus mosquitoes, but not those of Drosophila melanogaster or Spodoptera frugiperda (fall armyworm). Infectious cDNA clones of the genomic RNAs of FHV, NoV and PaV allow the initiation of their respective replicative cycle in many cell types on transfection of plasmid DNA or in vitro transcripts.
Transmission
NoV is transmissible to suckling mice by Aedes aegypti mosquitoes. It causes paralysis and death when injected into suckling mice and suckling hamsters, but no disease in adult animals. In their insect hosts, alphanodaviruses typically cause stunting, paralysis, and death.
Species demarcation criteria in the genus
The following criteria can be applied to the demarcation of species within the Alphanodavirus genus:
- Biological properties (host range, vectors, mode of transmission). Since the natural host ranges of the nodaviruses have generally not been examined in detail but may in some cases be broad, virus isolation from a new host is not, in itself, evidence of a new nodavirus species.
- Antigenic properties. Antisera raised against different isolates or strains of a single nodavirus species should exhibit high levels of cross-reactivity in Western blot and/or neutralization analyses. Lower levels of cross-reactivity in these assays using antisera against all previously recognized nodavirus species can provide evidence of a novel nodavirus.
- Virion physical/physicochemical characteristics.
- Virion electrophoretic mobility. Intact virus particles migrate with characteristic electrophoretic mobilities in non-denaturing agarose gel, so virion mobility should be compared with those of other nodavirus species.
- Sedimentation coefficient, buoyant density. Virion sedimentation coefficient and buoyant density should be compared with those of other nodavirus species.
- Structural protein characteristics. The electrophoretic mobilities in SDS-PAGE of the CP precursor or its cleavage products should be compared with those of other nodavirus species.
- Genome molecular characteristics.
- RNA electrophoretic mobilities. In the absence of sequence information, the electrophoretic mobilities of the viral genomic RNAs should be compared with those of other nodaviruses.
- RNA hybridization properties. In the absence of differences in RNA electrophoretic mobilities, the molecular hybridization properties of the viral genomic RNAs should be compared with those of other nodaviruses.
- Genome sequence characteristics. The nt sequence of the two genomic RNAs should be compared with those of other nodaviruses. Because the nodavirus genome is segmented, reassortment can occur and the two genome segments may have distinct evolutionary lineages.
Application of these criteria: In practice, while the five criteria above may be suggestive of a new species, definitive demarcation is based on the nt sequence of the viral CP gene. The two closest recognized species are BBV and FHV, whose RNA2 sequences show 80% identity at the nt level and 87% identity at the aa sequence level. Their RNA1 sequences, however, are 99% identical.
List of species in the genus Alphanodavirus
| Black beetle virus |
|
|
| Black beetle virus | RNA1[X02396=NC_001411] | (BBV) |
|
| RNA2[X00956=NC_002037] |
|
| Boolarra virus |
|
|
| Boolarra virus | RNA1[AF329080=NC_004142] | (BoV) |
|
| RNA2[X15960=NC_004145] |
|
| Flock House virus |
|
|
| Flock House virus | RNA1[X77156=NC_004146] | (FHV) |
|
| RNA2[X15959=NC_004144] |
|
| Nodamura virus |
|
|
| Nodamura virus | RNA1[AF174533=NC_002690] | (NoV) |
|
| RNA2[AF174534=NC_002691] |
|
| Pariacoto virus |
|
|
| Pariacoto virus | RNA1[AF171942=NC_003691] | (PaV) |
|
| RNA2[AF171943=NC_003692] |
|
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Alphanodavirus but have not been approved as species
| Drosophila line 1 virus | (DLV) |
| Gypsy moth virus | (GMV) |
| Lymantria ninayi virus Greenwood | (LNV) |
| Manawatu virus | (MwV) |
| New Zealand virus | (NZV) |
Genus Betanodavirus
Type species Striped jack nervous necrosis virus
Virion properties
Morphology
Virions are non-enveloped, spherical in shape, 25–30 nm in diameter, and have icosahedral symmetry (T=3). Distinct surface protrusions are observed by electron microscopy of negatively stained preparations (Figure 3). Image reconstruction of virus-like particles of Malabaricus grouper nervous necrosis virus (MGNNV) indicates that the CP of betanodaviruses has a two domain structure compared to the single domain structure of the CP of alphanodaviruses. The average diameter of the particle is 37 nm. In contrast with most alphanodaviruses, empty particles have been seen by electron microscopy of some preparations of betanodaviruses.
Physicochemical and physical properties
Virion buoyant density in CsCl of striped jack nervous necrosis virus (SJNNV) has not been reported but that of Dicentrarchus labrax encephalitis virus (DlEV) is about 1.31–1.36 g cm−3. Virions of DlEV are stable between pH 2 and 9 and resistant to heating at 56 °C for 30 min. Infectivity is resistant to extraction of virions with chloroform.
Nucleic acid
The genome consists of two molecules of positive sense ssRNA: RNA1 (Mr 1.01×106, 3.1 kb) and RNA2 (Mr 0.49×106, 1.4 kb). Both RNA molecules are encapsidated in the same virus particle, and both are required for infectivity. Both molecules are capped at their 5′ ends and lack poly(A) tails at their 3′ ends.
Proteins
Betanodavirus capsids contain 180 copies of a single structural protein of 42 kDa. In contrast to alphanodaviruses, maturation cleavage of this protein is not observed.
Lipids
None.
Carbohydrates
None.
Genome organization and replication
The betanodaviruses replicate in the cytoplasm. Infected cells contain three ssRNAs: RNA1 (Mr 1.01×106; 3.1 kb); RNA2 (Mr 0.49×106; 1.4 kb) and a sgRNA3 (Mr about 0.13×106; 0.4 kb) derived from RNA1. RNA3 is not packaged into virions. RNA1 encodes protein 1a (110 kDa), the RdRp. RNA2 encodes protein 2a (42 kDa), the CP. SJNNV RNA3 encodes protein B2 and has a potent RNA silencing-suppression activity, as in alphanodaviruses.
Antigenic properties
Betanodaviruses are cross-reactive by immunoblot analysis using polyclonal antisera but differential reactivity is observed with monoclonal antibodies. Virus neutralization with polyclonal antisera divides four betanodaviruses into three serotypes; serotype A for SJNNV, serotype B for TPNNV, and serotype C for RGNNV and BFNNV.
Biological properties
Host range
Nature: All species of the betanodaviruses were isolated from larvae, juvenile or adult marine fish, in which they cause “viral nervous necrosis” or “viral encephalopathy and retinopathy” associated with behavioral abnormalities and high mortalities. SJNNV and TPNNV have a limited host range: striped jack for SJNNV and tiger puffer for TPNNV. In contrast, RGNNV and BFNNV have a wide range of host fish; RGNNV is isolated from warm-water fishes and BFNNV is isolated from cold-water fishes. These diseases cause significant problems for the marine aquaculture industry.
Laboratory: Betanodaviruses replicate in cultured cells from striped snakehead fish (SNN-1 and E-11) and other cells derived from fish; GF-1 and others from groupers, SBL from sea bass, TV-1 from turbot, and SAF-1 from gilthead sea bream. A low level of virus replication is observed in mammalian (COS-1 and HeLa) cells.
Transmission: Betanodavirus antigens and/or CP genes are detected in eggs, larvae and ovaries of some inapparently infected hatchery-reared fish species, and the CP gene is frequently detected in a variety of wild fishes without any disease signs, indicating both horizontal and vertical modes of transmission of the virus.
Species demarcation criteria in the genus
The species demarcation criteria applied above for the alphanodaviruses also apply to betanodaviruses.
List of species in the genus Betanodavirus
| Barfin flounder nervous necrosis virus |
|
|
| Barfin flounder nervous necrosis virus - BF93Hok | RNA1[EU826137 = NC_011063] RNA2[EU826138 = NC_011064] | (BFNNV- BF93Hok) |
| Redspotted grouper nervous necrosis virus |
|
|
| Redspotted grouper nervous necrosis virus - SGWak97 | RNA1[AY324869 = NC_008040] RNA2[AY324870 = NC_008041] | (RGNNV- SGWak97) |
| Striped jack nervous necrosis virus |
|
|
| Striped jack nervous necrosis virus | RNA1[AB056571 = NC_003448] RNA2[AB056572 = NC_003449] | (SJNNV) |
| Tiger puffer nervous necrosis virus |
|
|
| Tiger puffer nervous necrosis virus - TPKag93 | RNA1[EU236148 = NC_013640] RNA2[EU236149 = NC_013641] | (TPNNV - TPKag93) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Betanodavirus but have not been approved as species
| Atlantic cod nervous necrosis virus | RNA1[EF433472] RNA2[EF433468] | (ACNNV) |
| Atlantic halibut nodavirus | RNA1[AJ401165] RNA2[AJ245641] | (AHNV) |
| Dicentrarchus labrax encephalitis virus | RNA2[U39876] | (DlEV) |
| Dragon grouper nervous necrosis virus | RNA1[AY721616] RNA2[AF245004, AY721615] | (DGNNV) |
| Greasy grouper nervous necrosis virus | RNA1[AF319555] RNA2[AF318942] | (GGNNV) |
| Japanese flounder nervous necrosis virus | RNA1[FJ748760] RNA2[D38527] | (JFNNV) |
| Lates calcarifer encephalitis virus | RNA2[EF591371] | (LcEV) |
| Malabaricus grouper nervous necrosis virus | RNA2[AF245003] | (MGNNV) |
| Seabass nervous necrosis virus | RNA2[Y08700] | (SBNNV) |
| Solea senegalensis nervous necrosis virus | RNA1[FJ803911] RNA2[AJ698113] | (SSNNV) |
| Turbot nodavirus | RNA2[AJ608266] | (TNV) |
List of unassigned species in the family Nodaviridae
None reported.
List of other related viruses which may be members of the family Nodaviridae but have not been approved as species
| Macrobrachium rosenbergii nodavirus | RNA1[AY231436] RNA2[AY222840] | (MrNV) |
| Penaeus vannamei nodavirus | RNA2[EF137180] | (PvNV) |
| Wuhan nodavirus | RNA1[AY962576] RNA2[DQ233638] | (WhNV) |
Phylogenetic relationships within the family
Within the alphanodaviruses, CP sequences are 44–87% identical to one another at the aa level, whereas within the betanodaviruses, CP aa sequence identities are 80% or greater. The phylogenetic relationship between betanodavirus species and the viruses not assigned to any species has not yet been rigorously defined. However, the sequences of RNA2 of RGNNV, GGNNV, MGNNV and DGNNV are >95% identical at both the nt and aa level suggesting that future reclassification of some of these as strains of current betanodavirus species may be possible. The CP aa sequences of the alphanodaviruses are only about 10% identical to those of the betanodaviruses, insufficient to indicate common ancestry.
Similarity with other taxa
The omegatetraviruses such as Nudaurelia capensis omega virus (NωV) and Helicoverpa armigera stunt virus (HaSV) contain bipartite ssRNA genomes, but their RNAs are about twice the size of nodavirus RNAs and they have no 3’-terminal blockage. Tetraviruses also have larger capsids with T=4 icosahedral symmetry.
Derivation of names
The name of the family Nodaviridae is derived from that of the type species of the alphanodaviruses, Nodamura virus, named in turn after its place of isolation in Japan. By convention, the members of the alphanodavirus genus are named after their places of isolation (BoV, FHV, NoV and PaV) or the host from which they were isolated (BBV from the black beetle, Heteronychus arator). Betanodaviruses are pathogens of fish causing “viral nervous necrosis” and the members of the betanodavirus genus are therefore named after the host fish from which they were isolated, followed by NNV (nervous necrosis virus).
Further reading
Ball and Johnson, 1998 L.A. Ball, K.L. Johnson, L.K. Miller, L.A. Ball, Nodaviruses of insectsThe Insect Viruses. In: L.K. Miller, L.A. Ball, The Insect Viruses. Plenum, New York1998225–267.
Iwamoto et al., 2001 T. Iwamoto, K. Mise, K. Mori, M. Arimoto, T. Nakai, T. Okuno, Establishment of an infectious RNA transcription system for striped jack nervous necrosis virus, the Type Species of the Betanodavirus. J. Gen. Virol. 82 (2001) 2653–2662.
Johnson and Reddy, 1998 J.E. Johnson, V. Reddy, L.K. Miller, L.A. Ball, Structural studies of noda and tetravirusesThe Insect Viruses. In: L.K. Miller, L.A. Ball, The Insect Viruses. Plenum, New York1998171–223.
Johnson et al., 2001 K.N. Johnson, K.L. Johnson, R. Dasgupta, T. Gratsch, L.A. Ball, Comparisons among the larger genome segments of six nodaviruses and their encoded RNA replicases. J. Gen. Virol. 82 (2001) 1855–1866.
Munday et al., 2002 B.L. Munday, J. Kwang, N. Moody, Betanodavirus infections of teleost fish: a review. J. Fish Dis. 25 (2002) 127–142.
Thiéry et al., 2004 R. Thiéry, J. Cozien, C. de Boisseson, S. Kerbart-Boscher, L. Nevarez, Genomic classification of new betanodavirus isolates by phylogenetic analysis of the coat protein gene suggests a low host-fish species specificity. J. Gen. Virol. 85 (2004) 3079–3087.
Contributed by
Thiéry, R., Johnson, K.L., Nakai, T., Schneemann, A., Bonami, J.R. and Lightner, D.V.
Figures
Figure 1 (Left) Image reconstruction of a particle of Flock House virus (FHV). (Center) Schematic representation of a T=3 icosahedral lattice. (Right) Cryo-electron micrograph of particles of FHV; the bar represents 50 nm.
(Courtesy of N. Olson and T. Baker.)
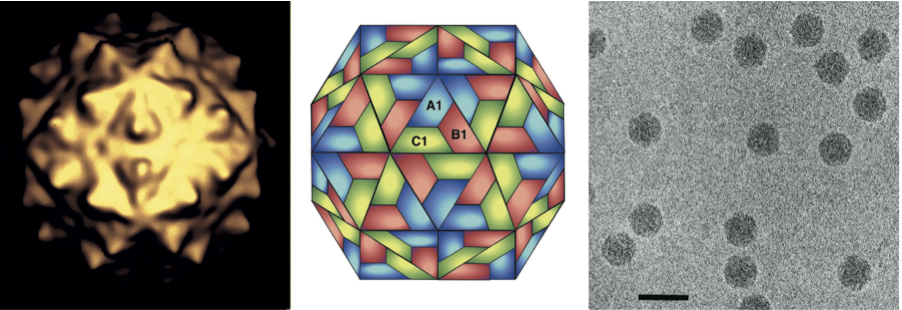
Figure 2 Alphanodavirus (Flock House virus; FHV) genome organization and strategy of replication.
(Adapted from L.A. Ball and K.L. Johnson.)

Figure 3 (Left) Image reconstruction of virus-like particles of Malabaricus grouper nervous necrosis virus (MGNNV) generated in Spodoptera frugiperda cells from a recombinant baculovirus expressing the MGNNV coat protein gene. (Center) Schematic representation of a T=3 icosahedral lattice. (Right) Cryo-electron micrograph of virus-like particles of MGNNV; the bar represents 40 nm.
(Courtesy of L. Tang and J.E. Johnson.)

