Family: Luteoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are 25 to 30 nm in diameter, hexagonal in outline and have no envelope (Figure 1). Amino acid sequence homology modelling using the X-ray crystal structure of rice yellow mottle virus (genus Sobemovirus) as a guide suggests that particles have 180 subunits arranged in T=3 icosahedra. Particles are composed of two CPs that encapsidate a genomic single stranded RNA.
Physicochemical and physical properties
Virion Mr is 5.6–6.0×106; buoyant density in CsCl is 1.39–1.42 g cm−3; S20,w is 106–127S. Virions are moderately stable and are insensitive to treatment with chloroform or non-ionic detergents, but are disrupted by prolonged treatment with high concentrations of salts. Luteovirus and polerovirus particles are insensitive to freezing.
Nucleic acid
Virions contain a single molecule of infectious, linear, positive sense ssRNA. The genome size is fairly uniform ranging from 5.6 kb to 6.0 kb. The RNAs do not have a 3′-terminal poly(A) tract. A small protein (VPg) is covalently linked to the 5′ end of the genomic RNAs of poleroviruses and the one enamovirus. The 5′ termini of barley yellow dwarf virus-PAV (BYDV-PAV) genomic RNA can be phosphorylated after treatment with alkaline phosphatase suggesting that the 5′ termini of luteovirus genomic RNAs are phosphorylated.
Proteins
There is a single major CP of 21–23 kDa encoded by ORF3 and smaller amounts of a “readthrough” protein, which is a fusion of the products of ORF3 and that of the contiguous ORF5. The readthrough protein may be associated with aphid transmission and/or virus particle stability.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Genomic RNAs of members of the Luteoviridae contain five or six ORFs that are predicted to encode proteins of between 4 and 132 kDa (Table 1; Figure 2). In poleroviruses and by extension the enamovirus, ORF0 (not present in members of the genus Luteovirus) encodes a symptom and host range determinant that functions as a suppressor of RNA silencing. Where present, it overlaps with ORF1. ORFs 1 and 2 overlap and encode the replication-related proteins. The major CP is encoded by ORF3, which is followed in frame by ORF5. The product of ORF4 (overlapping completely with ORF3, but absent from enaomovirus genomic RNA) has been shown to be required for long-distance movement of some luteoviruses and poleroviruses. Some luteovirus and polerovirus genomes contain an ORF 6, predicted to encode a small (≤6 kDa) protein, but no functions have been assigned to the proteins. Genera can be distinguished on the basis of the arrangements and sizes of the ORFs. Replication-related proteins encoded by ORFs 1 and 2 of the luteoviruses are not homologous to products of the corresponding ORFs of poleroviruses and the enamovirus. They are most similar to those of viruses in the family Tombusviridae, while the products of ORFs 1 and 2 of the poleroviruses and the enamovirus are related to those of sobemoviruses.
The differences among luteoviruses, poleroviruses and the enamovirus are principally in the 5′ end of the genome. ORFs 0, 1 and 2 are translated from the genomic RNA. ORF2 is translated by frameshift from ORF1 and thus shares an amino terminus with the product of ORF1. Polerovirus and enamovirus VPgs are cleaved from the products of ORF1 by upstream serine protease domains. ORFs 3, 4 (luteoviruses and poleroviruses) and 5 are expressed from a subgenomic RNA (sgRNA). ORF1 (poleroviruses and the enamovirus) and ORF4 (luteoviruses and poleroviruses) are expressed by leaky scanning. ORF5 is translated via a readthrough of the termination codon at the end of ORF3. Luteoviruses produce one or two additional sgRNAs, the larger of which from BYDV-PAV contains ORF6. Some poleroviruses produce additional sgRNAs.
There are no data on post-translational modification. Particles of some strains of cereal yellow dwarf virus-RPV (CYDV-RPV) contain 322 nt satellite RNAs. Virions of some isolates that consist of pea enation mosaic virus-1 (PEMV-1) together with the umbravirus, pea enation mosaic virus-2 (PEMV-2), contain 717-nt satellite RNAs in addition to genomic RNAs.
Table 1 Proteins of the different ORFs with sizes (kDa) and possible function(s)
| ORF | Luteovirus | Polerovirus | Enamovirus | Function of product |
| 0 | NA | 28–30 | 34 | Suppressor of RNA silencing |
| 1 | 39–42 | 66–72 | 84 | Helicase motifs in luteoviruses; protease and VPg in polero- and enamoviruses |
| 1+2 | 99–103 | 116–121 | 132 | RNA-dependent RNA polymerase |
| 3 | 22 | 22–23 | 21 | Major coat protein |
| 4 | 16–21 | 17–21 | NA | Probable MP |
| 3+5 | 72–80 | 67–80 | 55 | Minor coat protein expressed as C-terminal fusion to core coat protein; possible aphid transmission and virus particle stability factor |
| 6 | 4–7 | 7–9 | NA | Unknown |
Antigenic properties
Luteovirus and polerovirus particles are strongly immunogenic. Species within a genus are more closely related serologically than are species in different genera. Serological relationships may be detected when comparing disrupted virus particles that are not detectable when intact virions are tested. In gel diffusion assays, aphid-transmissible isolates sometimes display antigenic determinants that are absent from aphid-non-transmissible isolates. No serological relationships have been reported between enamoviruses and either luteoviruses or poleroviruses.
Biological properties
Host range
Several members of the family Luteoviridae have host ranges largely restricted to one plant family. For example, BYDVs and CYDVs infect several species in the family Poaceae, bean leafroll virus (BLRV) and soybean dwarf virus (SbDV) infect mainly legumes, and carrot red leaf virus infects mainly plants in the family Umbelliferae. Other members of the family Luteoviridae infect plants in several or many different families. For example, beet western yellows virus (BWYV) infects more than 150 species of plants in over 20 families.
Geographic distribution
Members of the family Luteoviridae have been reported from Arctic, temperate, sub-tropical, and tropical regions. Some of the viruses are found worldwide, such as BYDV, BWYV and potato leafroll virus (PLRV). Others have more restricted distributions, such as tobacco necrotic dwarf virus, which has been reported only from Japan, and groundnut rosette assistor virus, which has been reported from south Saharan countries in Africa.
Transmission
Transmission is in a circulative, non-propagative manner by specific aphid vectors. Viruses are acquired by phloem feeding, enter the hemocoel of the aphid via the hindgut (e.g., BYDV-PAV) or posterior midgut (e.g., PLRV) by a receptor mediated transport process, circulate in the hemolymph and enter the accessory salivary gland by a second receptor mediated transport event. Inoculation results from introduction of viruliferous saliva into the phloem tissues via the salivary duct during aphid feeding. PEMV-1 is readily transmitted mechanically, a property dependent on its multiplication in cells co-infected with PEMV-2 (Umbravirus).
Cytopathology
Luteovirus and polerovirus particles are largely confined to phloem cells; PEMV-1, with PEMV-2, is found in phloem and mesophyll tissues. Virus particles are found in both the nuclei and cytoplasm of infected cells. Luteoviruses and poleroviruses often cause phloem necrosis that spreads from inoculated sieve elements and causes symptoms by inhibiting translocation, slowing plant growth and inducing the loss of chlorophyll, which results in characteristic yellowing and dwarfing of infected plants.
Genus Luteovirus
Type species Barley yellow dwarf virus-PAV
Distinguishing features
Genome properties are the key features. There is no ORF0 and frameshift from ORF1 into ORF2 occurs at the termination codon of ORF1, and ORF1 and ORF2 overlap by less than 20 nt. ORF1 and ORF2 encode replication-related proteins that are most similar to those of viruses in the family Tombusviridae. The length of the non-coding sequence between ORF2 and ORF3 is about 100 nt. There is no evidence for the presence of a 5′ genome-linked protein. ORF4 is present and contained within ORF3. ORF5 is greater than 1350 nt in length.
Virion properties
Physicochemical and physical properties
Virion buoyant density in CsCl is 1.39–1.40 g cm−3; S20,w is 106–118S.
Nucleic acid
Sizes of positive sense ssRNA genomes are between 5,677 nt for BYDV-PAV and 5,964 nt for BLRV.
Genome organization and replication
In addition to the characters listed under distinguishing features, viruses within the genus produce two or three subgenomic RNAs from minus-strand templates, the largest of which expresses ORFs 3-5. The 3′-noncoding region contains a transcription enhancer that interacts with the 5′-nontranslated regions of genomic and large subgenomic RNAs to effect cap independent translation initiation.
Species demarcation criteria in the genus
Criteria used to demarcate species of the genus include:
- Differences in breadth and specificity of host range
- Failure of cross-protection in either one-way or two-way relationships
- Differences in serological specificity with discriminatory polyclonal or monoclonal antibodies
- Differences in amino acid sequence identity of any gene product of greater than 10%.
List of species in the genus Luteovirus
| Barley yellow dwarf virus-MAV |
|
|
| Barley yellow dwarf virus-MAV - PS1 | [D01213=NC_003680] | (BYDV-MAV-PS1) |
| Barley yellow dwarf virus-PAS |
|
|
| Barley yellow dwarf virus-PAS - 129 | [AF218798=NC_002160] | (BYDV-PAS-129) |
| Barley yellow dwarf virus-PAV |
|
|
| (Barley yellow dwarf virus-rgv=rice giallume) |
|
|
| Barley yellow dwarf virus-PAV - Australia | [X07653=NC_004750] | (BYDV-PAV-AUS) |
| Bean leafroll virus |
|
|
| (Legume yellows virus) |
|
|
| (Michigan alfalfa virus) |
|
|
| (Pea leafroll virus) |
|
|
| Bean leafroll virus - Michigan | [AF441393=NC_003369] | (BLRV-MI) |
| Rose spring dwarf-associated virus |
|
|
| Rose spring dwarf-associated virus - California | [EU024678=NC_010806] | (RSDaV-CA) |
| Soybean dwarf virus |
|
|
| (Subterranean clover red leaf virus) |
|
|
| Soybean dwarf virus - Tas-1 | [L24049=NC_003056] | (SbDV-Tas-1) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Luteovirus but have not been approved as species
| Barley yellow dwarf virus-GAV | [AY220739=NC_004666] | (BYDV-GAV) |
Genus Polerovirus
Type species Potato leafroll virus
Distinguishing features
Polerovirus genomic RNAs have VPgs linked to their 5′ termini and possess an ORF0 and a non-coding region between ORF2 and ORF3 of about 200 nt. ORF1 and ORF2 encode replication-related proteins, which are most similar to those of sobemoviruses. The frameshift from ORF1 into ORF2 occurs upstream of the termination of ORF1, and ORFs 1 and 2 overlap by more than 400 nt. Polerovirus genomes differ from those of the enamovirus in that ORF4 is present within ORF3 and ORF5 is greater than 1200 nt.
Virion properties
Physicochemical and physical properties
Virion buoyant density in CsCl is 1.39–1.42 g cm−3; S20,w is 115–127S.
Nucleic acid
Sizes of ssRNA genomes are between 5,641 nt for turnip yellows virus and 5,987 nt for PLRV.
Genome organization and replication
See “Distinguishing features” above.
Species demarcation criteria in the genus
See criteria for the genus Luteovirus.
List of species in the genus Polerovirus
| Beet chlorosis virus |
|
|
| Beet chlorosis virus - 2a | [AF352024=NC_002766] | (BChV-2a) |
| Beet mild yellowing virus |
|
|
| Beet mild yellowing virus - 2ITB | [X83110=NC_003491] | (BMYV-2ITB) |
| Beet western yellows virus |
|
|
| (Malva yellows virus) |
|
|
| (Turnip mild yellows virus) |
|
|
| Beet western yellows virus - USA | [AF473561=NC_004756] | (BWYV-US) |
| Carrot red leaf virus |
|
|
| Carrot red leaf virus - UK1 | [AY695933=NC_006265] | (CtLRV-UK1) |
| Cereal yellow dwarf virus-RPS |
|
|
| Cereal yellow dwarf virus-RPS - Mex1 | [AF235168=NC_002198] | (CYDV-RPS-Mex1) |
| Cereal yellow dwarf virus-RPV |
|
|
| Cereal yellow dwarf virus-RPV - NY | [L25299=NC_004751] | (CYDV-RPV-NY) |
| Chickpea chlorotic stunt virus |
|
|
| Chickpea chlorotic stunt virus - Et-fb-am1 | [AY956384=NC_008249] | (CpCSV-Et-fb-am1) |
| Cucurbit aphid-borne yellows virus |
|
|
| Cucurbit aphid-borne yellows virus - N | [X76931=NC_003688] | (CABYV-N) |
| Melon aphid-borne yellows virus |
|
|
| Melon aphid-borne yellows virus - Beijing | [EU000534=NC_010809] | (MABYV-BJ) |
| Potato leafroll virus |
|
|
| (Solanum yellows virus) |
|
|
| (Tomato yellow top virus) |
|
|
| Potato leafroll virus - UK:Scotland | [D00530=NC_001747] | (PLRV-UK) |
| Sugarcane yellow leaf virus |
|
|
| Sugarcane yellow leaf virus - Florida | [AF157029=NC_000874] | (ScYLV-FL) |
| Tobacco vein distorting virus |
|
|
| Tobacco vein distorting virus - China:Longlin | [EF529624=NC_010732] | (TVDV-CN) |
| Turnip yellows virus |
|
|
| Turnip yellows virus - FL-1 | [X13063=NC_003743] | (TuYV-FL1) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Polerovirus but have not been approved as species
| Cotton leafroll dwarf virus | [GQ379224*] | (CLRDV) |
| Suakwa aphid-borne yellows virus | [FJ425878*] | (SABYV) |
* Sequences do not comprise the complete genome.
Genus Enamovirus
Type species Pea enation mosaic virus-1
Distinguishing features
Enamovirus (PEMV-1) genomic RNA contains an ORF0, but does not contain an ORF4 (present in luteoviruses and poleroviruses). The non-coding intergenic region between ORF2 and ORF3 is about 200 nt in length. ORF1 and ORF2 encode replication-related proteins that are most similar to those of sobemoviruses. Frameshift from ORF1 into ORF2 occurs upstream of the termination of ORF1, and ORF1 and ORF2 overlap by more than 400 nt. The PEMV-1 genome contains an ORF5 of about 900 nt.
Virion properties
Physicochemical and physical properties
Enamovirus virions have a Mr of about 5.6×106, buoyant densities in CsCl of 1.42 g cm−3, and S20,w of 107–122S.
Genome organization and replication
Genomic RNA of PEMV-1 is 5,706 nt and has a 5′ VPg.
Antigenic properties
Virions produced in plants infected with PEMV-1 together with PEMV-2 (Umbravirus) are moderately antigenic.
Biological properties
PEMV-1 occurs as part of a complex with PEMV-2 (Umbravirus) and induces mosaic symptoms and enations. Unlike other members of the family Luteoviridae, PEMV-1 is readily transmitted mechanically, a property dependent on its multiplication in cells co-infected with PEMV-2. Aphid transmissibility is conferred by PEMV-1, but can be lost after several mechanical passages. Virions are found in mesophyll tissue as well as in vascular tissue. The genome of PEMV-1 is capable of autonomous replication in protoplasts, but is dependent on PEMV-2 to support systemic invasion.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Enamovirus
| Pea enation mosaic virus-1 |
|
|
| Pea enation mosaic virus-1 - WSG | [L04573=NC_003629] | (PEMV-1-WSG) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Enamovirus but have not been approved as species
None reported.
List of unassigned species in the family Luteoviridae
| Barley yellow dwarf virus-GPV |
|
|
| Barley yellow dwarf virus-GPV - 04FX6 | [EF174408*] | (BYDV-GPV-04FX6) |
| Wheat yellow dwarf virus-RPV | [FM865413=NC_012931] | (WYDV-RPV) |
| Barley yellow dwarf virus-RMV |
|
|
| Barley yellow dwarf virus-RMV - Illinois | [Z14123*] | (BYDV-RMV-IL) |
| Barley yellow dwarf virus-bv |
|
|
| Barley yellow dwarf virus-SGV - NY | [AY541038*] | (BYDV-SGV-NY) |
| Chickpea stunt disease associated virus |
|
|
| Chickpea stunt disease associated virus - IC | [Y11530*] | (CpSDaV-IC) |
| Groundnut rosette assistor virus |
|
|
| Groundnut rosette assistor virus - M16GCP | [AF195824*] | (GRAV-M16GCP) |
| Indonesian soybean dwarf virus |
|
|
| Indonesian soybean dwarf virus - IND |
| (ISDV-IND) |
| Sweet potato leaf speckling virus |
|
|
| Sweet potato leaf speckling virus - Peru | [DQ655700*] | (SPLSV-Peru) |
| Tobacco necrotic dwarf virus |
|
|
| Tobacco necrotic dwarf virus - Japan |
| (TNDV-JA) |
Species names are in italic script; names of isolates, strains and synonyms are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the family Luteoviridae but have not been approved as species
| Chickpea yellows virus | [GQ118150*] | (CpYV) |
| Lentil stunt virus | [GQ118152*] | (LSV) |
* Sequences do not comprise the complete genome.
Phylogenetic relationships within the family
The three genera within the family Luteoviridae share very similar structural protein genes (ORFs 3 and 5) whose products show varying levels of serological relatedness. Phylogenetic analysis of the predicted amino acid sequences of the polymerases (ORF2) clearly separate the members of the family Luteoviridae into the three genera (Figure 3).
The nucleotide sequences of BLRV and SbDV (genus Luteovirus) lack ORF0, like those of luteoviruses, and the predicted amino acid sequences of their replication proteins are similar to those of the luteoviruses. However, their structural proteins are more closely related to those of poleroviruses (Figure 3). Conversely, sugarcane yellow leaf virus (genus Polerovirus) contains an ORF0 and its ORFs 1 and 2 are most closely related to those of other poleroviruses, whereas ORFs 3 and 4 are most closely related to those of the luteoviruses and ORF5 is most closely related to the readthrough protein gene of the enamovirus. These viruses may be recombinants between the genera.
Similarity with other taxa
Viruses in the family Luteoviridae have replication-related and structural proteins that are sufficiently similar to those in other genera to suggest evolutionary relationships. The putative luteovirus polymerases resemble those of members of the family Tombusviridae. In contrast, polymerases of poleroviruses and enamoviruses resemble those of viruses in the genus Sobemovirus. These polymerase types are thought to be very distant in evolutionary terms. The CP amino acid sequences of PLRV and rice yellow mottle virus, a sobemovirus, share 33% similarity, which has been used to predict the structure of PLRV and other members of the family Luteoviridae. It has been suggested that the genomes of the Luteoviridae originated by recombination between ancestral genomes containing the structural protein genes characteristic of the family Luteoviridae and genomes containing either of the two polymerase types.
Derivation of names
Enamo: from pea enation mosaic virus.
Luteo: from Latin luteus, “yellow”.
Polero: from potato leaf roll virus.
Further reading
Demler et al., 1995 S.A. Demler, G.A. De Zoeten, G. Adam, K.F. Harris, B.D. Harrison, A.F. Murant, Pea enation mosaic enamovirus: properties and aphid transmissionThe Plant Viruses 5: Polyhedral virions and bipartite RNA genomes. In: B.D. Harrison, A.F. Murant, The Plant Viruses 5: Polyhedral virions and bipartite RNA genomes. Plenum, New York and London1995303–344.
Gray and Gildow, 2003 S. Gray, F.E. Gildow, Luteovirus-aphid interactions. Annu. Rev. Phytopathol. 41 (2003) 539–566.
Hauser et al., 2000 S. Hauser, M. Stevens, C. Mougel, H.G. Smith, C. Fritsch, E. Herrbach, O. Lemaire, Biological, serological, and molecular variability suggest three distinct polerovirus species infecting beet or rape. Phytopathol. 90 (2000) 460–466.
Martin and D’Arcy, 1990 R.R. Martin, C.J. D’Arcy, Relationships among luteoviruses based on nucleic acid hybridization and serological studies. Intervirol. 31 (1990) 23–30.
Mayo and Ziegler-Graff, 1996 M.A. Mayo, V. Ziegler-Graff, Molecular biology of luteoviruses. Adv. Virus Res. 46 (1996) 413–460.
Miller et al., 2002 W.A. Miller, S.J. Liu, R. Beckett, Barley yellow dwarf virus: Luteoviridae or Tombusviridae?. Mol. Plant Pathol. 3 (2002) 177–183.
Miller and White, 2006 W.A. Miller, K.A. White, Long-distance RNA-RNA interactions in plant virus gene expression and replication. Annu. Rev. Phytopathol. 44 (2006) 447–467.
Stevens et al., 2005 M. Stevens, B. Freeman, H.Y. Liu, E. Herrbach, O. Lemaire, Beet poleroviruses: close, friends or distant relatives?. Mol. Plant Pathol. 6 (2005) 1–9.
Taliansky et al., 2003 M. Taliansky, M.A. Mayo, H. Barker, Potato leafroll virus: a classic pathogen shows some new tricks. Mol. Plant Pathol. 4 (2003) 81–89.
Terradot et al., 2001 L. Terradot, M. Souchet, V. Tran, D.G. Ducray-Bourdin, Analysis of a three-dimensional structure of Potato leafroll virus coat protein obtained by homology modelling. Virology. 286 (2001) 72–82.
Contributed by
Domier, L.L.
Figures
Figure 1 (Left) Diagram of the proposed structure of luteovirus particles. (Center) Negative contrast electron micrograph of particles of barley yellow dwarf virus-PAV (BYDV-PAV) and (right) pea enation mosaic virus-1 (PEMV-1), isolated by means of sucrose density gradient centrifugation and stained with uranyl acetate. Bars represent 100 nm.
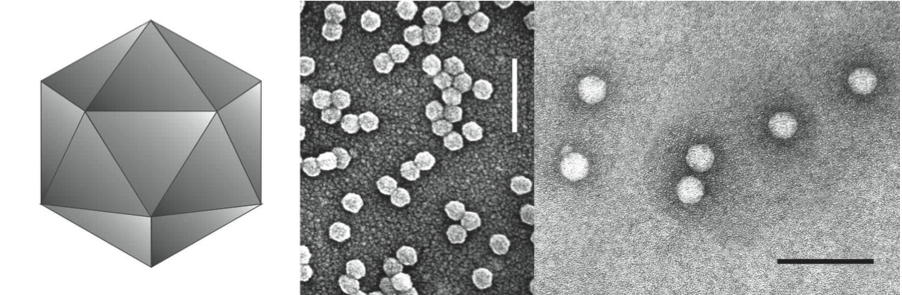
Figure 2 Diagram of the genome organization and map of the translation products typical of viruses in each genus of the family Luteoviridae. Solid lines represent RNA; boxes represent ORFs; thinner boxes represent translation products; grey ovals represent VPgs.
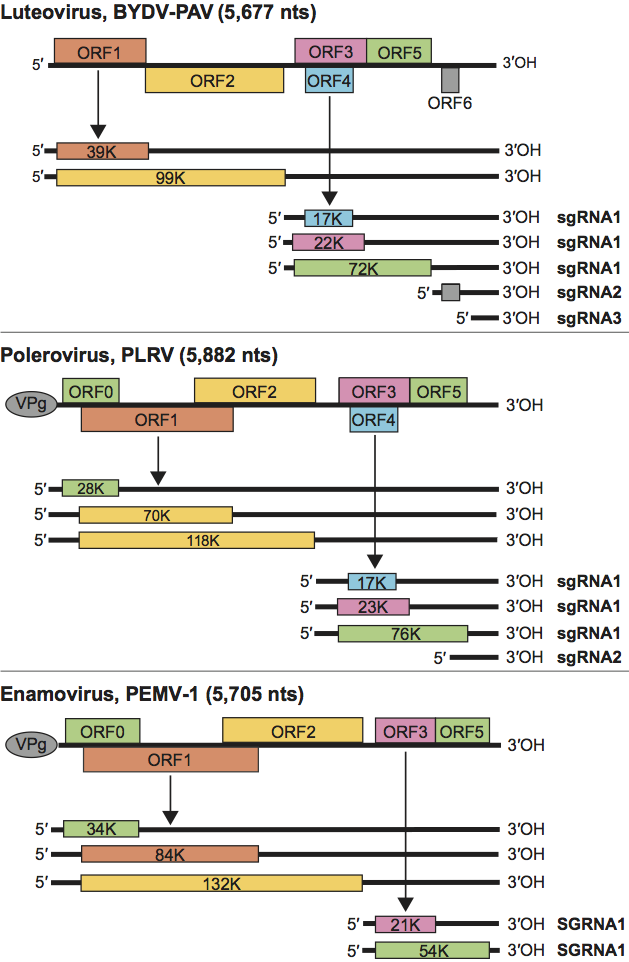
Figure 3 Phylogenetic analyses of the (left) readthrough protein (ORFs 3 and 5) and (right) polymerase (ORF2) sequences of representatives of species in the family Luteoviridae. Amino acid sequences were aligned with CLUSTALX and neighbour-joining trees constructed with MEGA 4. Bootstrap values above 50% are indicated.

