Family: Leviviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are spherical and exhibit icosahedral symmetry (T=3) with a diameter of about 26 nm. There is no envelope (Figure 1).
Physicochemical and physical properties
Virion Mr varies from 3.6 to 4.2×106 depending on the genus. The S20,w value is 80–84S and buoyant density in CsCl is 1.46 g cm−3. Infectivity is ether-, chloroform- and low-pH-resistant, but is sensitive to RNase and detergents. Inactivation by UV light and chemicals is comparable to that of other icosahedral viruses containing ssRNA.
Nucleic acid
Virions contain one molecule of positive sense ssRNA of 3466–4276 nt: size and gene arrangement vary with genus. RNA makes up 39% of the virion weight. The 5′ nucleotide carries a triphosphate, while at the 3′ terminus a non-templated A residue is added by the replicase (see Figures 2, 4).
Proteins
The capsid contains 180 copies of CP (14 kDa) arranged in an icosahedron. The structure of the protein shell of several ssRNA phages has been solved by X-ray crystallography, and shows 60 quasi-symmetric AB- and 30 symmetric CC′-dimers. The A and C subunits are situated around the three-fold axes, and the B subunits around the five-fold axes of the icosahedron. CP has no structural similarity to those of eukaryote icosahedral RNA viruses. The X-ray structure of the capsid in a complex with the 19 nt operator shows interaction of the dimers with this hairpin. Operators found in the various phages are shown in Figure 3.
Each virion contains one copy of the A-protein (35–61 kDa), which is required for maturation of the virion and for pilus attachment (Figure 1). Alloleviviruses also contain several copies of the readthrough protein in their capsid.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Members of the family Leviviridae that propagate in E. coli infect by adsorption to the sides of F(ertility) pili. (Non-coliphages such as Pseudomonas phage PP7 (PP7) and Acinetobacter phage AP205 (AP205) bind to other pili.) This event leads to cleavage of the A-protein and release of the RNA from the virion into the bacterium. The infecting RNA encodes a replicase, which assembles with three host proteins (ribosomal protein S1 and translation elongation factors EF-Tu and EF-Ts) to form the active RNA polymerase. A fourth protein, called Host Factor, not associated with the polymerase complex but acting directly on the RNA, is needed for synthesis of the minus strand. Members of the two genera use different Host Factors. Plus-strand synthesis requires, besides the virus-coded replicase, only EF-Tu and EF-Ts as cofactors. Late in infection coat-protein dimers act as translational repressors of the replicase gene by binding to an RNA hairpin, the operator that contains the start site of this gene. This protein–RNA complex is considered to also be the nucleation site for encapsidation. Virions assemble in the cytoplasm around phage RNA. It is unknown at which point the A-protein (and readthrough protein) is assembled in the virion but it is assumed to be an early step since the A-protein cannot be incorporated into preformed virions lacking the protein. Infection usually results in cell lysis releasing thousands of phages per cell. The lysis protein short-circuits the membrane potential and somehow activates the bacterial autolysins leading to degradation of the peptidoglycan network.
Antigenic properties
Members of the family Leviviridae are highly antigenic.
Biological properties
Members of the family Leviviridae occur worldwide and are abundantly present in sewage, waste water, animal and human faeces. In Asia a particular geographic distribution has been noticed with respect to the four levivirus species. It has also been proposed that the various species have a preference for particular hosts, e.g. members of Enterobacteria phage Qbeta are found predominantly in human waste. The evidence is not conclusive. RNA bacteriophages are harmless for humans. Members of the family Leviviridae not only infect enterobacteria but also species of the genera Caulobacter, Pseudomonas and Acinetobacter and probably many other Gram-negative bacteria, provided they express the appropriate pili on their surface. RNA coliphages are often used as indicators for the presence of enteroviruses in waste and surface water. There is renewed interest in phage therapy to combat bacterial infections.
Genus Levivirus
Type species Enterobacteria phage MS2
Distinguishing features
Leviviruses contain the short version of the genome and have a separate gene for cell lysis, which partly overlaps the replicase coding region in the +1 reading frame (see Figure 2). Overlap with the CP gene is variable. Genome size ranges from 3466 for GA (Enterobacteria phage BZ13) to 3577 for fr (Enterobacteria phage MS2) (Figure 2). Leviviruses and alloleviviruses use different Host Factors for their polymerase holoenzyme. The levivirus Host Factor has been isolated but has not been genetically identified. Generally, the replicases from leviviruses poorly replicate allolevivirus RNA and vice versa.
The sequence of RNA of AP205, an RNA phage growing on Acinetobacter tentatively identified its lysis gene in the unusual location of the 5′ end. The absence of a readthrough protein was taken as criterion to classify AP205 as a levivirus (Figure 4).
Recently, the genome sequence of PRR1 (3,573 nt), a phage with a broad host range, was determined; its genetic map is identical to that of the leviviruses.
Genome organization and replication
Figure 2 shows the map of the levivirus genome. Lysis and replicase synthesis are dependent on translation of the CP gene: early CP nonsense mutants are deficient in replicase and lysis protein synthesis. Translational starts at the lysis gene were shown to be reinitiations by ribosomes that had completed CP-gene reading but had not yet detached themselves from the message. A small fraction of these ribosomes manages to back up to the lysis start. Part of the replicase ribosome binding site is base-paired to an upstream sequence located in the coat coding region. A ribosome translating the CP cistron disrupts this interaction, thereby exposing the replicase start site (when not blocked by a CP dimer, which is the case late in infection). The CP gene is freely accessible to ribosomes.
Maturation or A-protein is translated from a specific RNA folding intermediate which has an accessible ribosome-binding site. This intermediate exists for a short time on nascent strands. Full-length RNA reaches an equilibrium folding in which the start site of the A-protein gene is inaccessible. It is believed that the purpose of these control mechanisms is to facilitate the switch from translation of the viral RNA to its replication. One of the binding sites of the replicase holoenzyme is the start of the CP gene. Binding of the enzyme to this site squeezes out ribosomes from CP, lysis and replicase genes. At this stage the A-protein gene is folded in its ribosome-inaccessible state and replication can proceed without interference from translation.
The polymerase of GA has been purified, that of MS2 may be unstable. Except for the Host Factor the polymerases of leviviruses and alloleviviruses contain the same subunits.
Antigenic properties
Antigenic specificity is distinct from that of members of the genus Allolevivirus.
Species demarcation criteria in the genus
A major difference between members of the species Enterobacteria phage MS2 and Enterobacteria phage BZ13 (formerly called subgroups I and II) is a deletion of about 60 nt in the 3′-UTR of members of Enterobacteria phage BZ13, comprising three small RNA hairpins (Figure 5). There is also a 35 nt deletion in the replicase gene of members of Enterobacteria phage BZ13 producing a shorter hairpin stem. Furthermore, the percentage of aa or nt sequence identity is dramatically lower between the two species than between strains within a species. Species can also be distinguished by serological means and by species-specific antisense DNA probes.
List of species in the genus Levivirus
| Enterobacteria phage BZ13 |
|
|
| Enterobacteria phage GA | [X03869] | (GA) |
| Enterobacteria phage JP34 | [J04343] | (JP34) |
| Enterobacteria phage KU1 | [AF227250] | (KU1) |
| Enterobacteria phage TH1 | [AB218930] | (TH1) |
| Enterobacteria phage BZ13 | [FJ483839] | (BZ13) |
| Enterobacteria phage MS2 |
|
|
| Enterobacteria phage MS2 | [V00642] | (MS2) |
| Enterobacteria phage f2 |
| (f2) |
| Enterobacteria phage fr | [X15031] | (fr) |
| Enterobacteria phage JP501 | [AF227251] | (JP501) |
| Enterobacteria phage M12 | [AF195778] | (M12) |
| Enterobacteria phage R17 | [EF108465] | (R17) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Levivirus but have not been approved as species
| Acinetobacter phage AP205 | [AF334111] | (AP205) |
| Pseudomonas phage PP7 | [X80191] | (PP7) |
| Pseudomonas phage PRR1 | [DQ836063] | (PRR1) |
Genus Allolevivirus
Type species Enterobacteria phage Qbeta
Distinguishing features
Alloleviviruses contain the longer version of the genome (Figure 2). The extra RNA encodes a C-terminal extension of CP arising by occasional suppression of the CP gene termination codon. The readthrough protein is present at about 12 copies per virion. Together with the A-protein, it is necessary for infection. Its precise role is not known. There is no separate lysis gene. Cell lysis is a secondary function of the A-protein. Genome length varies between 4217 nt for Qβ (Enterobacteria phage Qbeta) and 4276 nt for SP (Enterobacteria phage F1) (Figure 2).
Genome organization and replication
Genome organization is shown Figure 2. The RNA polymerase of Qβ has been purified and the enzyme can amplify Qβ RNA in vitro. The crystal structure of the enzyme has been solved. The Host Factor has been purified and genetically characterized. It is the product of the hfq gene. In the uninfected cell the protein functions in the transition to stationary phase. In particular, it stimulates translation of the mRNA encoding the σ38 protein involved in transcription of stationary phase genes. Hfq is a sequence non-specific ssRNA binding protein with some preference for A-residues. It is heat-resistant and acts as a pentamer. The protein helps the polymerase to get access to the 3′ end of the plus strand, which exists in a base-paired and therefore inactive state. In Figure 6 the secondary structure of the 3′UTR of Qβ RNA is shown; the 3′-terminal 6 nt are taken up in long-distance interaction with ld IX.
Although the polymerases are specific for their own RNA, the interaction with RNA involves host-encoded subunits (EF-Tu, S1 and Hfq) that have no sequence specificity. An important contribution to template activity is provided by the higher order structure of Qβ RNA (Figure 6). For instance, destroying two out of the eight base pairs that make up the central pseudoknot in Qβ RNA, here indicated as ld X, lowers replication 100-fold. The higher order structures of the RNAs of phages PP7 (not allocated to a current species) and SP (Enterobacteria phage F1) are shown in Figure 7.
The switch from translation to replication is as in leviviruses and was first formulated for Qβ. Control of the maturation protein is slightly different. The time window for producing the A-protein is not set by the lifetime of a folding intermediate, as for MS2, but by the time it takes the polymerase to move from about position 60, the start of the A-protein gene, to about position 470 where the complement to the Shine–Dalgarno sequence of the A-protein gene is located. Once this complement is synthesized, pairing between the two regions blocks furthers translation.
Antigenic properties
Antigenic specificity is distinct from that of members of the genus Levivirus.
Species demarcation criteria in the genus
The major difference between Enterobacteria phage Qbeta and Enterobacteria phage F1 (formerly called subgroups III and IV respectively) is a deletion of about 90 nt in the maturation-protein gene of Qβ, corresponding to a bifurcated hairpin. There is also the extra stem-loop (V1) in the 3′UTR of members of Enterobacteria phage Qbeta that is lacking in members of Enterobacteria phage F1. Species can also be differentiated by serological criteria and by species-specific antisense DNA probes. Finally, the percentage of aa or nt sequence identity is dramatically lower between the two species than between strains within a species.
List of species in the genus Allolevivirus
| Enterobacteria phage F1 |
|
|
| Enterobacteria phage F1 | [EF068134] | (F1) |
| Enterobacteria phage ID2 |
| (ID2) |
| Enterobacteria phage NL95 | [AF059243] | (NL95) |
| Enterobacteria phage SP | [X07489] | (SP) |
| Enterobacteria phage TW28 |
| (TW28) |
| Enterobacteria phage Qbeta |
|
|
| Enterobacteria phage Qβ | [AY099114] | (Qβ) |
| Enterobacteria phage M11 | [AF052431] | (M11) |
| Enterobacteria phage MX1 | [AF059242] | (MX1) |
| Enterobacteria phage ST |
| (ST) |
| Enterobacteria phage TW18 | [FJ483840] | (TW18) |
| Enterobacteria phage VK | [EU372698] | (VK) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Allolevivirus but have not been approved as species
None reported.
List of other related viruses which may be members of the family Leviviridae but have not been approved as species
| Caulobacter phage Cb2 |
| (Cb2) |
| Caulobacter phage Cb4 |
| (Cb4) |
| Caulobacter phage Cb5 | [HM066936] | (Cb5) |
| Caulobacter phage Cb8r |
| (Cb8r) |
| Caulobacter phage Cb9 |
| (Cb9) |
| Caulobacter phage Cb12r |
| (Cb12r) |
| Caulobacter phage Cb23r |
| (Cb23r) |
| Caulobacter phage CP2 |
| (CP2) |
| Caulobacter phage CP18 |
| (CP18) |
| Caulobacter phage Cr14 |
| (Cr14) |
| Caulobacter phage Cr28 |
| (Cr28) |
| Enterobacteria phage B6 |
| (B6) |
| Enterobacteria phage B7 |
| (B7) |
| Enterobacteria phage C-1 |
| (C-1) |
| Enterobacteria phage C2 |
| (C2) |
| Enterobacteria phage fcan |
| (fcan) |
| Enterobacteria phage Folac |
| (Folac) |
| Enterobacteria phage Iα |
| (Iα) |
| Enterobacteria phage M |
| (M) |
| Enterobacteria phage pilHα |
| (pilHα) |
| Enterobacteria phage R23 |
| (R23) |
| Enterobacteria phage R34 |
| (R34) |
| Enterobacteria phage ZG/1 |
| (ZG/1) |
| Enterobacteria phage ZIK/1 |
| (ZIK/1) |
| Enterobacteria phage ZJ/1 |
| (ZJ/1) |
| Enterobacteria phage ZL/3 |
| (ZL/3) |
| Enterobacteria phage ZS/3 |
| (ZS/3) |
| Enterobacteria phage α15 |
| (α15) |
| Enterobacteria phage β |
| (β) |
| Enterobacteria phage μ2 |
| (μ2) |
| Enterobacteria phage τ |
| (τ) |
| (other enterobacteriophages, with many plasmid specificities, have been reported). | ||
| Pseudomonas phage 7s |
| (7s) |
Phylogenetic relationships within the family
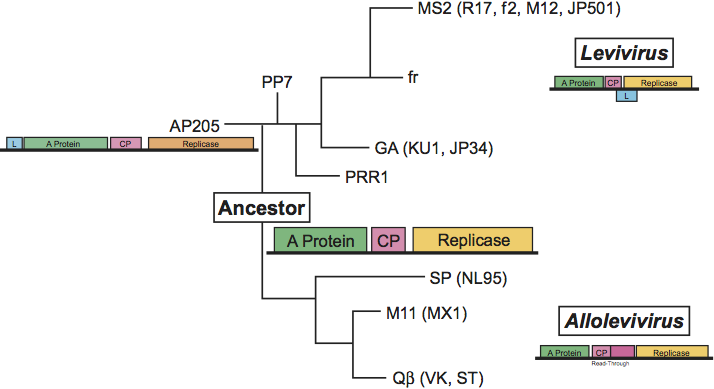
A tentative phylogenetic tree of the family Leviviridae is given in Figure 8. Relationships have been based first on deeply rooted features such as the genetic map and second on similarity in RNA folding, in particular the one present at the 3′UTR which is conserved in its outline. As a result there is a fundamental split between leviviruses and alloleviviruses because they have different maps. The two non-coli leviviruses AP205 and PP7 have been placed closer to the ancestor than the coli leviviruses because they have the same folding of their 3′UTR as the alloleviviruses (see Figure 7). As a result MS2 and GA are closer to the non-coliphages than to coliphage Qβ. PP7 is placed closer to MS2 than AP205 because AP205 has its lysis gene in a different position. PP7 has it in the same position as MS2, fr and GA. PRR1 is positioned between PP7 and the group II phages (GA) owing to the folding of its 3′UTR.
In this scheme, the ancestor contains only the three basic genes and the A-protein has the double function of lysis and maturation (infection). We assume that its 3′UTR is folded in the simple way of PP7 (AP205) and Qβ (SP) (see Figure 7).
The subdivision of each genus into two species is based on criteria explained above. Based on the sequence it is possible to make subtle distinctions between strains within a species. For example, MS2, R17, f2, M12 and JP501 are extremely close (ca. 95% identity) whereas fr is much further away (ca. 80% identity), has some features of members of Enterobacteria phage BZ13, but is still clearly a member of Enterobacteria phage MS2.
Similarity with other taxa
Not reported.
Derivation of names
Levi: from Latin levis, “light”.
Allo: from Greek allon, “other”.
Further reading
Ackermann and Dubow, 1987 . In: . CRC Press, Boca Raton, FL1987.
Furuse, 1987 K. Furuse, S.M. Goyal, C.P. Gerber, G. Bitton, Distribution of the coliphages in the environmentPhage Ecology. In: S.M. Goyal, C.P. Gerber, G. Bitton, Phage Ecology. John Wiley and Sons, New York198787–124.
Horn et al., 2006 W.T. Horn, K. Tars, E. Grahn, C. Helgstrand, A.J. Baron, H. Lago, C.J. Adams, D.S. Peabody, S.E. Phillips, N.J. Stonehouse, L. Liljas, P.G. Stockley, Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2. Structure. 14 (2006) 487–495.
Jacobson et al., 1998 A.B. Jacobson, R. Arora, M. Zuker, C. Priano, C.H. Liu, D.R. Mills, Structural plasticity in RNA and its role in the regulation of translation in Qβ. J. Mol. Biol. 275 (1998) 589–600.
Kidmose et al., 2010 R.T. Kidmose, N.N. Vasiliev, A.B. Chetverin, G.R. Andersen, C.R. Knudsen, Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins. Proc. Natl Acad. Sci., U S A. 107 (2010) 10884–10889.
Miranda et al., 1997 G. Miranda, D. Schuppli, I. Barrera, C. Hausherr, J.M. Sogo, H. Weber, Recognition of Qβ plus strand RNA as a template by Qβ replicase: role of RNA interactions mediated by ribosomal protein S1 and Host Factor. J. Mol. Biol. 267 (1997) 1089–1103.
Olsthoorn et al., 1995 R.C.L. Olsthoorn, G. Garde, T. Dayhuff, J.F. Atkins, J. Van Duin, Nucleotide sequence of a single-stranded RNA phage from Pseudomonas aeruginosa: kinship to coliphages and conservation of regulatory RNA structures. Virology. 206 (1995) 611–625.
Schuppli et al., 2000 D. Schuppli, J. Georgijevic, H. Weber, Synergism of mutations in Qβ RNA affecting host factor dependence of Qβ replicase. J. Mol. Biol. 295 (2000) 149–154.
van den Worm et al., 2006 S.H. van den Worm, R.I. Koning, H.J. Warmenhoven, H.K. Koerten, J. van Duin, Cryo electron microscopy reconstructions of the Leviviridae unveil the densest icosahedral RNA packing possible. J. Mol. Biol. 363 (2006) 858–865.
van Duin, 1988 J. van Duin, R. Calendar, Single stranded RNA bacteriophagesThe Bacteriophages. In: R. Calendar, The Bacteriophages. Plenum Press, New York1988117–167.
Contributed by
van Duin, J. and Olsthoorn, R.C.L.
Figures
Figure 1 (Left) Schematic representation of a levivirus: the RNA inside the virion is highly structured. (Upper right) Escherichia coli bacterium with Enterobacteria phage MS2 (MS2) particles attached to its F-pili (courtesy A.B. Jacobson). The inset is a -pilus with phage- enlargement. (Courtesy R.I. Koning and H.K. Koerten.) (Lower right) Image reconstruction obtained from cryo-electron microscopy of MS2 phages. View from outside (left) and inside (right).
(Courtesy R.I. Koning and H.K. Koerten.)
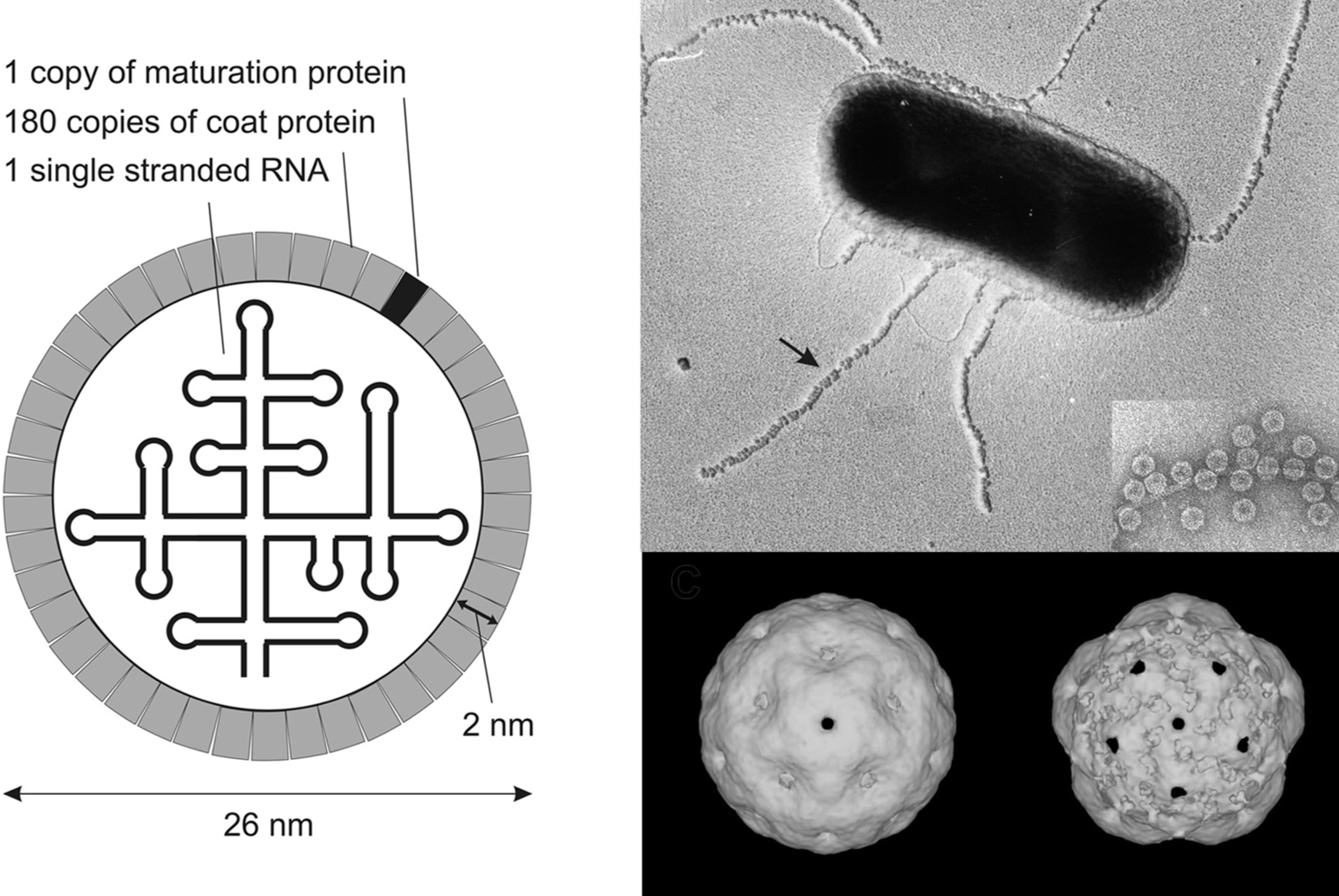
Figure 2 General genetic map of a representative levivirus Enterobacteria phage MS2 (MS2) and an allolevivirus Enterobacteria phage Q (Q). The maturation protein is also called A-protein. The lysis gene overlaps the replicase gene in a+1 frameshift. Arrows indicate repression of replicase translation by capsid protein binding to an RNA hairpin structure (the operator) present at the start of the gene. The UGA nonsense codon (nt 1743) is occasionally (ca. 6%) misread as tryptophan to produce the readthrough protein.
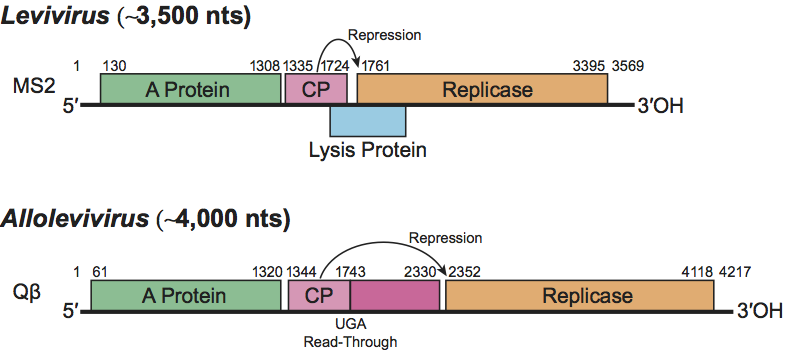
Figure 3 2D structures of operator hairpins in several RNA phages. Note the presence of a second, putative operator hairpin in all three phages. The structure shown for PRR1 has not been confirmed yet.
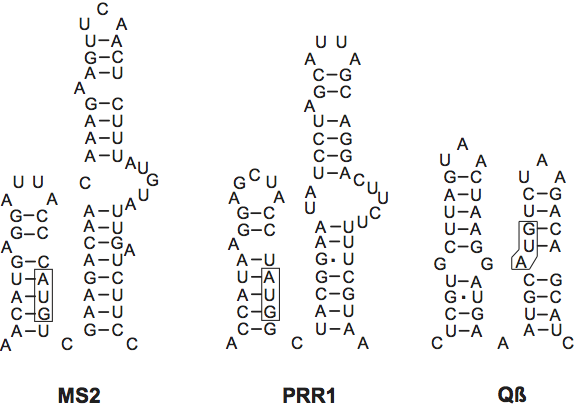
Figure 4 Genetic map of Acinetobacter phage AP205 (AP205). Note the location of the tentative lysis gene at the 5-terminus. AP205 is unusually long for a levivirus. This map corrects the one previously published (Klovins, J. et al. (2002). J. Gen. Virol. (2002), 83, 15231533). A: A-protein; CP: capsid protein; R: replicase; L: lysis.
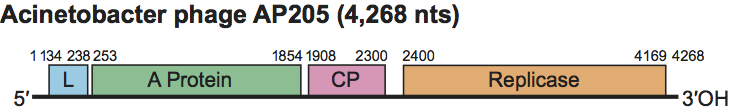
Figure 5 Comparison of the RNA folding in the 3UTR of Enterobacteria phage MS2 (MS2) and Enterobacteria phage GA (GA). GA lacks the three stem-loops U4, U5 and U6. In MS2 stem-loops V1 and V2 are part of the A-protein binding site. The other part of the proteins binding site is located around nt 400.
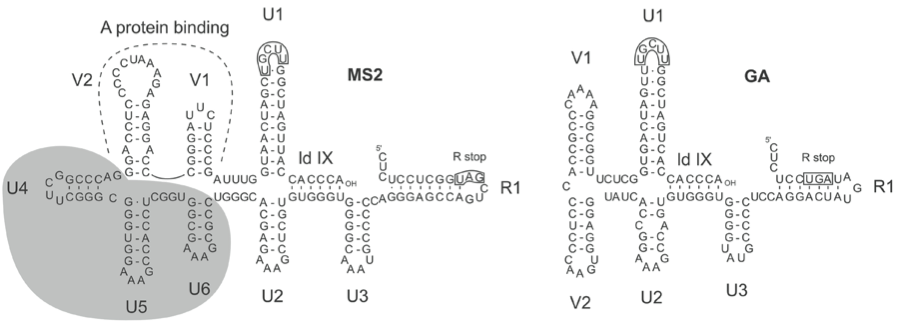
Figure 6 RNA secondary structure for Enterobacteria phage Q (Q) RNA from nt 2966 to the 3 end (nt 4217) marked as AOH. The UAA stop codon (nt 4119) of the replicase gene is boxed. Replicase Domain 2 (RD2) containing 1062 nt has been replaced by a dotted circle. Breaking two or three basepairs in the central pseudoknot (ldX) or ldVIII abolishes replication. However, breaking the pairs in ld IX, which buries the 3-terminal nucleotides, stimulates replication. Production of minus strand is also inhibited by deletion of stem-loops U1, V1, V2 or U2. (R1 and R2 were not tested.)
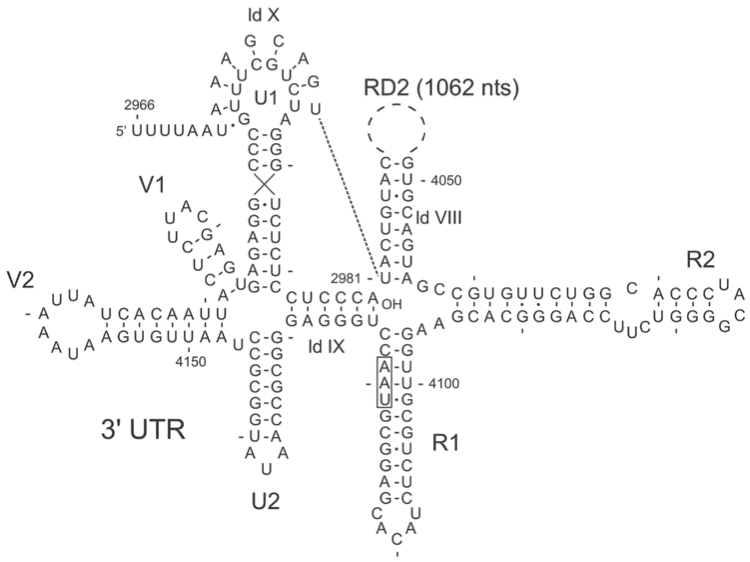
Figure 7 RNA secondary structure in the 3UTR of Pseudomonas phage PP7 (PP7) and Enterobacteria phage SP (SP). The folding of PP7 RNA is much more like that of SP RNA than that of either MS2 or GA (Figure 5). Compared to MS2 the stem-loops U3, U4, U5, U6 and one of the two V-loops are missing. The boxed sequence in the loop of hairpin U1 is conserved in all viruses of the family Leviviridae. The sequence is part of the central pseudoknot in Q. The pseudoknot is believed to exist also in the other phages.
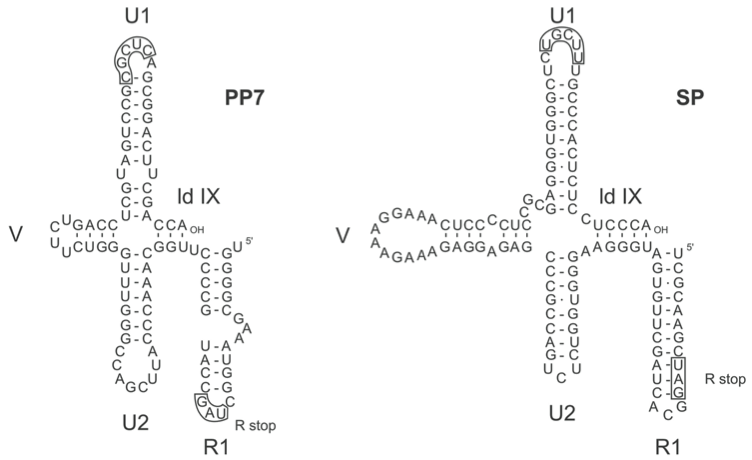
Figure 8 Proposed phylogenetic tree for the family Leviviridae. Distances are arbitrary. The ancestor only has the three basic genes. Lysis is effected by the A-protein as it still is today in Q. Presumably, fitness of the ancestor was restricted by the double function of the A-protein (Bollback, J.P. and Huelsenbeck, J.P. (2001). J. Mol. Evol., 52, 117128). The leviviruses solved the problem by evolving a separate lysis protein either encoded on a vacant region of the genome (AP205) or resulting from a ribosomal restart following translation termination at the end of the capsid gene (other leviviruses). Once restrictions on the A-protein were relaxed the gene could evolve in various directions to better fulfill its remaining function: virion maturation and infection. Two features of leviviruses can be explained by this scenario: first, lysis genes have variable startpoints (even between MS2 and fr or between GA and KU1) and secondly, of the three old genes, the A-protein gene shows the lowest sequence conservation. The alloleviviruses solved the dual-function problem by transferring part of the maturation and infection function to a new protein, readthrough, which arose by an insertion between coat and replicase genes. Presumably, this allowed A-protein to improve its lysis function. Such a scenario would provide a different reason why also in the alloleviviruses A-protein is least conserved of the old genes. Abbreviations of virus names are provided in the tables.