Genus: Idaeovirus
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Type species: Raspberry bushy dwarf virus
Distinguishing features
A monotypic genus for a pollen transmitted virus with isometric particles containing two genomic and one subgenomic positive sense, single stranded RNAs.
Virion properties
Morphology
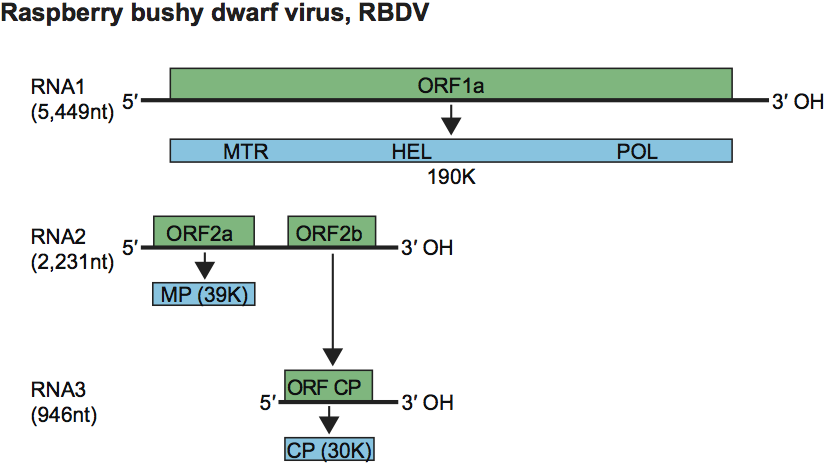
Virions are isometric, about 33 nm in diameter and are not enveloped. They appear flattened in electron micrographs of preparations negatively stained with uranyl salts.
Negative contrast electron micrograph of particles of an isolate of Raspberry bushy dwarf virus, stained with uranyl formate/sodium hydroxide. The bar represents 100 nm.
Physicochemical and physical properties
Virion Mr is about 7.5×106 (calculated from the S20,w of 115S). The buoyant density of aldehyde-fixed particles in CsCl is 1.37 g cm−3. Particles are readily disrupted in neutral chloride salts and by SDS.
Nucleic acid
Virion preparations contain three species of linear, positive sense, ssRNA of about 5.5 kb (RNA-1), 2.2 kb (RNA-2) and 1 kb (RNA-3). These RNA molecules are not polyadenylated.
Genome organization and replication
The genome is bipartite. RNA-1 has a single, major ORF encoding a 188 kDa protein that contains sequence motifs characteristic of viral RNA helicases and polymerases. RNA-2 has two ORFs: that in the 5′-terminal half encodes a protein of about 39 kDa which has some slight sequence similarities with cell-to-cell movement proteins of other viruses; that in the 3′-terminal half encodes the 30 kDa CP. RNA-2 is probably a template for the production of RNA-3, that comprises the 3′-most 946 nt of RNA-2 and is a sgRNA for CP expression. The 3′-terminal non-coding 18 nt of RNA-1 and RNA-2 (and hence of RNA-3) are the same and the 3′-terminal 70 nt can be arranged in similar extensively base-paired structures. Infected leaves contain dsRNA corresponding in size to double stranded forms of RNA-1 and RNA-2. In vitro translation of RNA isolated from purified virus particles yields three major proteins, of about 190, 44 and 31 kDa (CP), which are the translation products, respectively, of RNA-1, RNA-2 and RNA-3. Using infectious clones it has been shown that RNA1 and RNA2 together have low infectivity but that the addition of RNA3 (and probably also virus coat protein itself, although these experiments have not been done) greatly stimulates virus replication. This resembles the mechanism of “genome activation” that operates also with Alfalfa mosaic virus and ilarviruses.
Scale diagram of RNA species found in particles of raspberry bushy dwarf virus (RBDV). The open boxes represent the ORFs. The positions of putative domains are indicated as “MTR”, a methyltransferase motif, “HEL”, an NTP-binding motif, “POL”, an RNA-dependent RNA polymerase motif. MP signifies the putative movement protein role of the 39 kDa protein and CP is the coat protein.
Antigenic properties
Particles are moderate immunogens.
Biological properties
In nature, the host range was thought to be confined to Rubus species, all but one in the subgenus Idaeobatus. However, RBDV has recently been found causing an infection in grapevines. The experimental host range of RBDV is fairly wide. The virus occurs in all tissues of the plant, including seed and pollen, and is transmitted in association with pollen, both vertically to the seed and horizontally to the pollinated plant. This is the only known method of natural spread. Experimentally, the virus can be transmitted by mechanical inoculation. The virus occurs throughout the world wherever raspberry is grown. Infection of raspberry and blackberry is often symptomless but in some cultivars may be associated with “yellows disease” and/or “crumbly fruit”, a major economic problem in raspberry production. Confusingly, black raspberry necrosis virus is the main cause of bushy dwarf disease in Lloyd George raspberry. However, because the additional presence of RBDV in plants contributes significantly to the intensity of the disease symptoms usually observed in the field, it can also be regarded as an integral component of the disease syndrome.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Idaeovirus
| Raspberry bushy dwarf virus |
|
|
| Raspberry bushy dwarf virus-R15 | [S51557=NC_003739+S55890=NC_003740] | (RBDV-R15) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Idaeovirus but have not been approved as species
| Unnamed citrus idaeovirus | [DQ100358*] |
|
* Sequence does not comprise a complete genome segment.
Similarity with other taxa
RBDV resembles viruses of the genus Ilarvirus in having easily deformable particles; and the species Alfalfa mosaic virus as well as the ilarviruses (both family Bromoviridae) in being transmitted in association with pollen and in using the CP to activate genome replication. RBDV RNA-2 resembles RNA-3 of viruses in family Bromoviridae in the arrangement and sizes of its encoded gene products, the generation of a 3′-terminal sgRNA and in the structured nature of the 3′ ends of the molecules. The sequence of the translation product of RBDV RNA-1 resembles, in different parts, sequences in the translation products of viruses in the family Bromoviridae and to a lesser extent the sequence of the replicase protein (ca. 183 kDa) of tobamoviruses. Idaeoviruses, therefore, belong to the “alphavirus-like” supergroup.
Derivation of name
Idaeo: from idaeus, specific name of raspberry Rubus idaeus.
Further reading
Barnett, O.W. and Murant, A.F. (1970). Host range, properties and purification of raspberry bushy dwarf virus. Ann. Appl. Biol., 65, 435-449.
Jones, A.T., Mayo, M.A. and Murant, A.F. (1996). Raspberry bushy dwarf idaeovirus. In: Harrison, B.D. and Murant, A.F. (Eds.), The Plant Viruses, vol. 5: Polyhedral Virions and Bipartite RNA Genomes. Plenum Press, New York, pp. 283-301.
Jones, A.T., Murant, A.F., Jennings, D.L. and Wood, G.A. (1982). Association of raspberry bushy dwarf virus with raspberry yellows disease; reaction of Rubus species and cultivars, and the inheritance of resistance. Ann. Appl. Biol., 100, 135-147.
MacFarlane, S.A. and McGavin, W.J. (2009). Genome activation by Raspberry bushy dwarf virus coat protein. J. Gen. Virol., 90, 747-753.
Mayo, M.A., Jolly, C.A., Murant, A.F. and Raschke, J.H. (1991). Nucleotide sequence of raspberry bushy dwarf virus RNA-3. J. Gen. Virol., 72, 469-472.
Murant, A.F., Chambers, J. and Jones, A.T. (1974). Spread of raspberry bushy dwarf virus by pollination, its association with crumbly fruit, and problems of control. Ann. Appl. Biol., 77, 271-281.
Murant, A.F., Mayo, M.A. and Raschke, J.H. (1986). Some biochemical properties of raspberry bushy dwarf virus. Acta Hort., 186, 23-30.
Natsuaki, T., Mayo, M.A., Jolly, C.A. and Murant, A.F. (1991). Nucleotide sequence of raspberry bushy dwarf virus RNA-2: a bicistronic component of a bipartite genome. J. Gen. Virol., 72, 2183-2189.
Pleško, I.M., Marn, M.V., Širca, S. and Urek, G. (2009). Biological, serological and molecular characterization of Raspberry bushy dwarf virus from grapevine and its detection in the nematode Logidorus juvenilis. Eur. J. Plant Pathol., 123, 261-268.
Ziegler, A., Natsuaki, T., Mayo, M.A., Jolly, C.A. and Murant, A.F. (1992). Nucleotide sequence of raspberry bushy dwarf virus RNA-1. J. Gen. Virol., 73, 3213-3218.
Contributed by
MacFarlane, S.A.
Figures
Figure 1 Negative contrast electron micrograph of particles of an isolate of Raspberry bushy dwarf virus, stained with uranyl formate/sodium hydroxide. The bar represents 100 nm.
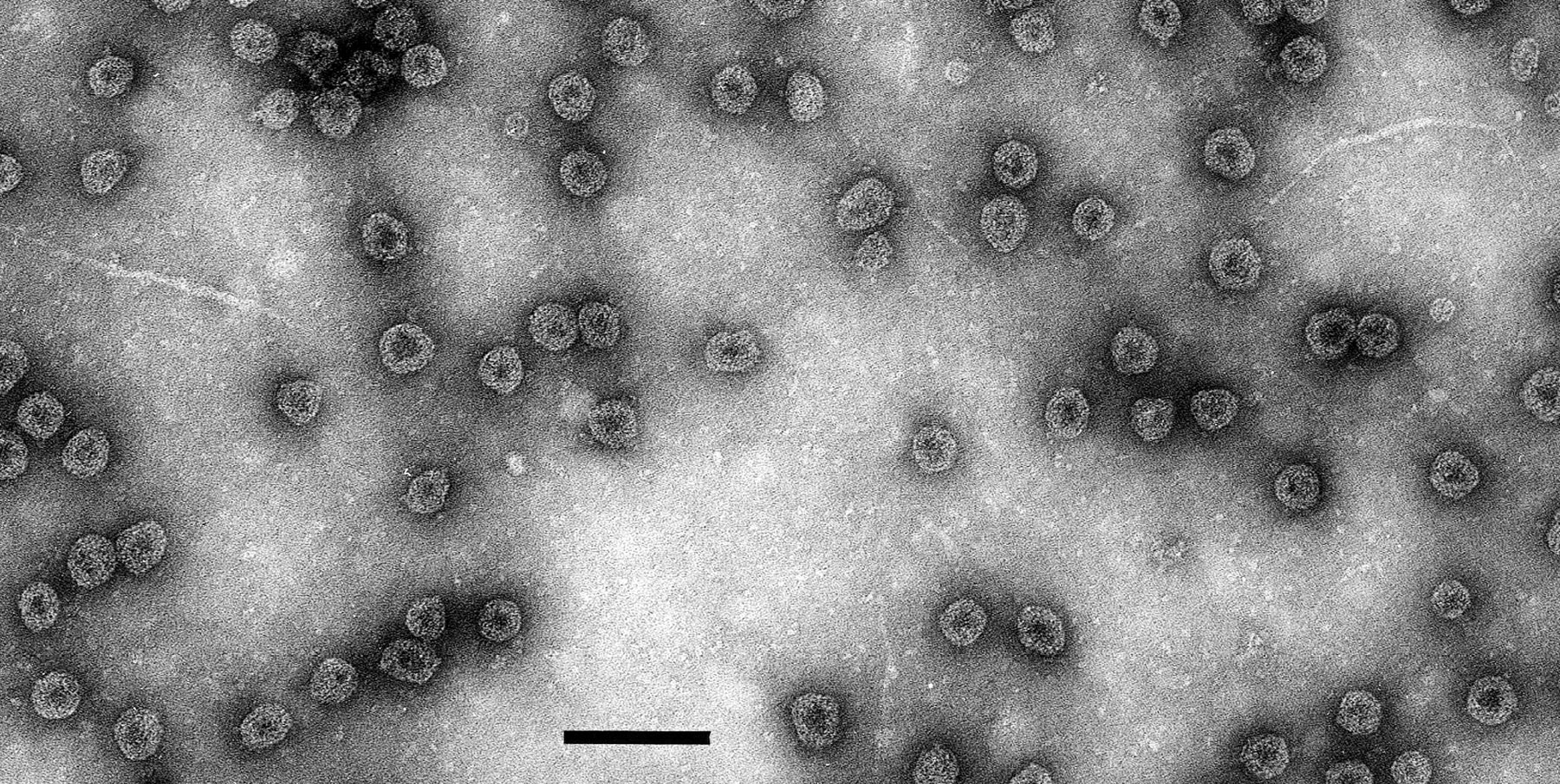
Figure 2 Scale diagram of RNA species found in particles of raspberry bushy dwarf virus (RBDV). The open boxes represent the ORFs. The positions of putative domains are indicated as MTR, a methyltransferase motif, HEL, an NTP-binding motif, POL, an RNA-dependent RNA polymerase motif. MP signifies the putative movement protein role of the 39 kDa protein and CP is the coat protein.