Family: Hypoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Hypovirus
Type species Cryphonectria hypovirus 1
Virion properties
Morphology
No true virions are associated with members of this family. Pleomorphic vesicles 50–80 nm in diameter, devoid of any detectable viral structural proteins but containing replicative form dsRNA and polymerase activity, are the only virus-associated particles that can be isolated from infected fungal tissue. (See Figure 1.)
Physicochemical and physical properties
Mr of vesicles is unknown. They have a buoyant density in CsCl of approximately 1.27–1.3 g cm−3 and sediment through sucrose as a broad component of approximately 200S. Their pH stability is unknown. The vesicles can be purified in pH 5.0 buffer and resuspended in pH 7.0 buffer. pH optimum for polymerase activity in vitro is 8.0; the optimum Mg++ concentration for polymerase activity is 5 mM. Activity decreases dramatically at pH less than 7.0 or more than 9.0. The vesicles are unstable when heated, or dispersed in lipid solvents. Optimal temperature for polymerase activity is 30 °C; temperatures over 40 °C inactivate polymerase activity. Deoxycholate at concentrations of more than 0.5% inactivates polymerase activity.
Nucleic acid
Vesicles contain linear dsRNA, 9–13 kbp in size, that of one isolate of Cryphonectria hypovirus 1, the type species, being 12,712 nt. Apparently only one strand is employed in mRNA transcription. The coding (positive) strand contains a short 3′-poly(A) tail, which is 20–30 nt in length when analyzed as a component of the dsRNA.
In the absence of an identifiable virion RNA, hypoviruses were originally classified, along with several other fungal viruses, as dsRNA viruses. However, the ability of positive sense ssRNA, not dsRNA, to initiate infection is consistent with their current classification among the positive strand RNA viruses. The predominant dsRNA form found in infected mycelia appears to be replicative intermediate or replicative form RNA. The accumulation of plus strand RNA, by contrast, is low due to the RNA silencing antiviral defense response of the fungal host, but greatly increases in fungal strains in which the corresponding dicer or argonaute genes have been disrupted. The presence of shorter-than-full-length, internally deleted, defective interfering (DI) replicative form dsRNA molecules is common among some members, and satellite-like dsRNAs are present in other members. The host RNA silencing pathway has been reported to promote DI RNA production. No function has been ascribed to any ancillary dsRNA. The 5′-terminus of the positive strand of dsRNA from Cryphonectria hypovirus 1 (CHV-1) is blocked, but the nature of the blocking group is unknown. The 5′-terminus of the negative strand is unblocked. Both 5′-termini of dsRNA from Cryphonectria hypovirus 3-GH2 (CHV-3/GH2) are unblocked.
Proteins
No structural proteins have been described for members of this family. Functions have been assigned to several nonstructural polypeptides encoded by members of the family. The 5′-proximal coding domain, ORFA, of CHV-1/EP713 RNA encodes a papain-like protease, p29, and a highly basic protein, p40, derived, respectively, from the N-terminus and C-terminus of polyprotein p69, by a p29-mediated cleavage event (Figure 2). A presumptive NS protein identified in vitro and in vivo, p29, has been shown by DNA-mediated transformation to contribute to suppression of host pigmentation, reduced sporulation, reduced laccase accumulation, to serve as a suppressor of host RNA silencing, and to promote RNA recombination of a co-infecting mycoreovirus. Protein p40 enhances viral RNA accumulation. RdRp activity is associated with isolated vesicles of CHV-1/EP713. The calculated size of the CHV-1 ORFB product, which contains putative RNA polymerase and helicase domains and a second papain-like protease, p48, is approximately 250×103 based on deduced aa sequence from cDNA clones, but no protein of that size has yet been isolated from vesicles. Smaller virus-encoded proteins have been identified in the vesicle-associated polymerase complex, suggesting extensive processing of replication proteins and that ORFB processing occurs in vivo. There are no known external viral proteins. The polymerase transcribes ssRNA molecules in vitro that correspond in size to full-length dsRNA. Approximately 90% of the polymerase products in vitro are of positive polarity. The p48 protein is required for initiation but not maintenance of viral RNA replication. CHV-3 and CHV-4 each contains a putative UDP glycosyltransferase-encoding domain that is not present in CHV-1 or CHV-2, although the specific function of this domain has not been examined.
Lipids
Host-derived lipids make up the vesicles that encapsulate the viral dsRNA.
Carbohydrates
Carbohydrates similar to those involved in fungal cell wall synthesis are associated with vesicles.
Genome organization and replication
A 5′-leader of approximately 300–500 nt, including several AUG triplets, precedes the AUG codon that initiates the first long ORF. The viral coding region may be expressed from a single long ORF, or may be divided into two ORFs. If two ORFs are present, the shorter, 5′-proximal ORF is designated ORFA. Its product may or may not be autocatalytically cleaved, depending on the virus. The UAA termination sequence at the end of ORFA is part of the pentanucleotide UAAUG in all members with two ORFs investigated to date. The AUG of the UAAUG pentanucleotide initiates the other long ORF, ORFB. The N-terminal product of ORFB is a papain-like cysteine protease that autocatalytically releases from the growing polypeptide chain (e.g., P48 for CHV-1/EP713 or P52 for CHV-2/NB58). No further processing in vitro has been demonstrated for the remaining 300×103 polypeptide from this ORF. Phylogenetic relatedness to members of the positive sense, ssRNA genus Potyvirus has been demonstrated by comparisons of protease, polymerase and helicase domains, although these domains are positioned differently in the two families.
Antigenic properties
No antibody has ever been raised from virus particle preparations. Anti-dsRNA antibodies were used to confirm the replicative form dsRNA constituent by immuno-electron microscopy. Antibodies directed against CHV1-EP713-encoded p29 were used to demonstrate association with the trans-Golgi network membranes. Antibodies directed against the conserved RNA polymerase domain of ORFB, expressed in bacteria, were used to identify an 87 kDa protein in a CHV-1/EP713 infected isolate.
Biological properties
Confirmed members infect the chestnut blight fungus, Cryphonectria parasitica. Confirmed members result in reduced virulence (hypovirulence) on chestnut trees and altered fungal morphology in culture, but many possible family members have little or no discernible effect on the fungal host. Some possible members infect other filamentous fungi, e.g., Sclerotinia sclerotiorum. Infection of fungal mycelium is known only through fusion, or anastomosis, of infected hyphae with uninfected hyphae. The transmission rate through asexual spores (conidia) varies from a few to close to 100%. Transmission through sexual spores (ascospores) is not known to occur. Transmission via cell-free extracts has not been demonstrated but transfection of protoplasts with full-length synthetic transcripts has been successful for hypoviruses CHV-1/EP713, CHV-1/Euro7 and CHV-1/EP721. Confirmed members have been identified throughout chestnut-growing areas of Europe, North America and Asia. DsRNA-containing vesicles have been associated with abnormal Golgi apparati in freeze-substituted thin sections. No nuclear or mitochondrial associations, nor virus-associated inclusions, have been noted.
Species demarcation criteria in the genus
Species are differentiated based on major differences in genetic organization or genome expression, as well as by major differences in nucleic acid sequence identity. Thus, CHV-1 differs from CHV-2 in the presence or absence, respectively, of a papain-like proteinase in ORFA. CHV-1 and CHV-2 isolates share less than 60% overall aa sequence identity with each other. CHV-3 and CHV-4 each contain a single ORF, but isolates of the two species share less than 50% overall sequence identity. Infection by CHV-1 isolates results in a white or near-white phenotype in the fungus; CHV-2 infection results in an orange-brown phenotype; CHV-3 and CHV-4 isolates have little effect on fungal pigment. Infection by members of any of the four species may reduce fungal virulence.
List of species in the genus Hypovirus
| Cryphonectria hypovirus 1 |
|
|
| Cryphonectria hypovirus 1 - EP713 | [M57938] | (CHV-1/EP713) |
| Cryphonectria hypovirus 1 - Euro7 | [AF082191] | (CHV-1/Euro7) |
| Cryphonectria hypovirus 1 - EP721 | [DQ861913] | (CHV-1/EP721) |
| Cryphonectria hypovirus 2 |
|
|
| Cryphonectria hypovirus 2 - NB58 | [L29010] | (CHV-2/NB58) |
| Cryphonectria hypovirus 3 |
|
|
| Cryphonectria hypovirus 3 - GH2 | [AF188515] | (CHV-3/GH2) |
| Cryphonectria hypovirus 4 |
|
|
| Cryphonectria hypovirus 4 - SR2 | [AY307099] | (CHV-4/SR2) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Hypovirus but have not been approved as species
None reported.
List of unassigned species in the family Hypoviridae
None reported.
Phylogenetic relationships within the family
The hierarchy of relationships among members of this family is unknown. Many strains and isolates have been identified by RNA hybridization, RFLP, or nt sequence analysis. Among the four species, CHV-1 and CHV-2 are more closely related to each other and CHV-3 and CHV-4 are more closely related to each other.
Similarity with other taxa
The possession of an undivided positive sense RNA genome, expressed via long polyprotein precursors, demonstrates an affinity with viruses of the picornavirus supergroup. Deduced aa sequences of polymerase, helicase and protease motifs of members of the family Hypoviridae suggest that their closest relatives are bymoviruses in the family Potyviridae.
Derivation of name
Hypo: from hypovirulence.
Further reading
Chen and Nuss, 1999 B. Chen, D.L. Nuss, Infectious cDNA clone of hypovirus CHV1-Euro7: a comparative virology approach to investigate virus-mediated hypovirulence of the chestnut blight fungus Cryphonectria parasitica. J. Virol. 73 (1999) 985–992.
Choi and Nuss, 1992 G.H. Choi, D.L. Nuss, Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science. 257 (1992) 800–803.
Dawe and Nuss, 2001 A.L. Dawe, D.L. Nuss, Hypoviruses and chestnut blight: Exploiting viruses to understand and modulate fungal pathogenesis. Annu. Rev. Genet. 35 (2001) 1–29.
Hillman et al., 1994 B.I. Hillman, B.T. Halpern, M.P. Brown, A viral dsRNA element of the chestnut blight fungus with a distinct genetic organization. Virology. 201 (1994) 241–250.
Hillman and Suzuki, 2004 B.I. Hillman, N. Suzuki, Viruses of the chestnut blight fungus. Adv. Virus. Res. 63 (2004) 423–473.
Lin et al., 2007 H. Lin, X. Lan, H. Liao, T.B. Parsley, D.L. Nuss, B. Chen, Genome sequence, full-length infectious cDNA clone, and mapping of viral double-stranded RNA accumulation determinant of hypovirus CHV1-EP721. J. Virol. 81 (2007) 1813–1820.
Linder-Basso et al., 2005 D. Linder-Basso, J.N. Dynek, B.I. Hillman, Genome analysis of Cryphonectria hypovirus 4, the most common hypovirus species in North America. Virology. 337 (2005) 192–203.
Nuss, 1992 D.L. Nuss, Biological control of chestnut blight: an example of virus-mediated attenuation of fungal pathogenesis. Microbiol. Rev. 56 (1992) 561–576.
Nuss, 2005 D.L. Nuss, Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3 (2005) 632–642.
Smart et al., 1999 C.D. Smart, W. Yuan, R. Foglia, D.L. Nuss, D.W. Fulbright, B.I. Hillman, Cryphonectria hypovirus 3, a virus species in the Family Hypoviridae with a single open reading frame. Virology. 265 (1999) 66–73.
Contributed by
Nuss, D.L. and Hillman, B.I.
Figures
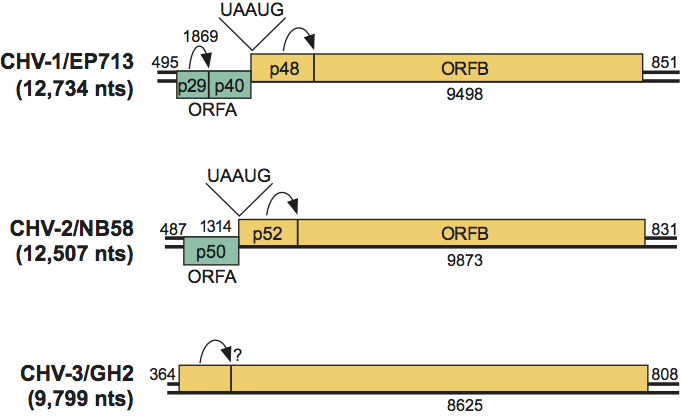
Figure 1 (Top) Thin section showing vesicles in fungal tissue; (bottom) thin section showing vesicle aggregate in fungal tissue surrounded by rough ER (from Newhouse et al. (1983). Can. J. Bot., 61, 389399). The bar represents 100 nm.
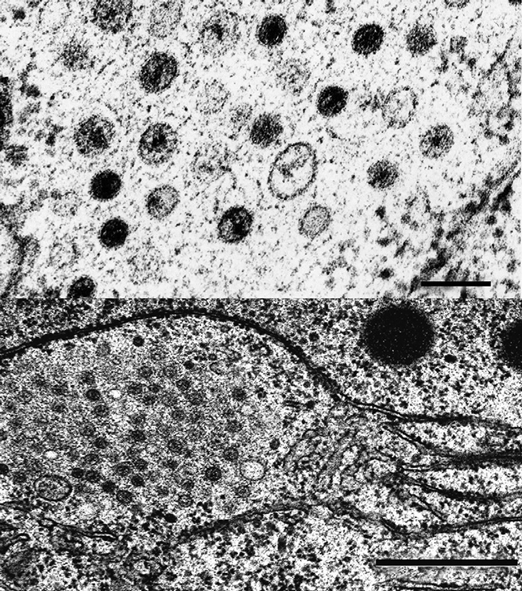
Figure 2 Genome organization of three members of the family Hypoviridae. Arrows represent known or suspected sites of autoproteolysis. Genome organization of Cryphonectria hypovirus 4 (CHV-4), which is 9.1 kb, is similar to Cryphonectria hypovirus 3 (CHV-3), but it is unknown whether CHV-4 undergoes autoproteolysis.