Family: Hepeviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions of hepatitis E virus (HEV) are icosahedral, non-enveloped, spherical particles with a diameter of approximately 27–34 nm (Figure 1). The capsid is formed by capsomeres consisting of homodimers of a single capsid protein forming the virus shell. Each capsid protein contains three linear domains forming distinct structural elements: S (the continuous capsid), P1 (three-fold protrusions), and P2 (two-fold spikes). Each domain contains a putative polysaccharide-binding site that may interact with cellular receptors. Native T=3 capsid contains flat dimers, with less curvature than those of T=1 virus-like particles (Figure 2).
Virion particles of avian hepatitis E virus (Figure 3) revealed by negative staining EM of bile samples from chickens with hepatitis–splenomegaly syndrome are similar in size and morphology to members of the genus Hepevirus.
Physicochemical and physical properties
Virion buoyant density is 1.35 to 1.40 g cm−3 in CsCl and 1.29 g cm−3 in glycerol and potassium tartrate gradients. Virion S20,w is 183S. Virion is sensitive to low-temperature storage (between −70 °C and +8 °C) and iodinated disinfectants. The virion of hepatitis E virus is more heat-labile than is hepatitis A virus: hepatitis E virus was about 50% inactivated at 56 °C and almost totally inactivated (96%) at 60 °C for 1 h. Liver suspensions containing avian hepatitis E virus remained infectious after treatment with chloroform and ether but lost infectivity after incubating at 56 °C for 1 h or 37 °C for 6 h. Viral infectivity in liver suspensions was reduced 1000-fold after treatment with 0.05% Tween-20, 0.1% NP40 and 0.05% formalin.
Nucleic acid
The HEV genome is a linear, positive sense, ssRNA molecule of approximately 7.2 kb, with a 5′-m7G cap structure and a 3′-poly(A) tail. The genome of avian hepatitis E virus is similar but only about 6.6 kb in size.
Proteins
Virions are constructed from a major capsid protein encoded by the second open reading frame (ORF2), and the CP may be proteolytically processed. A small immunoreactive protein (12.5 kDa) encoded by the third ORF (ORF3) has been identified and shown to exhibit multiple functions associated with virion morphogenesis and viral pathogenesis. Non-structural proteins encoded by the first major ORF (ORF1) have limited similarity with the “alpha-like supergroup” of viruses and contain domains consistent with a Mtr, RNA helicase, papain-like cysteine protease, and RdRp. The translational and posttranslational processes of the non-structural polyprotein remain unresolved. It remains unclear whether the non-structural polyprotein functions as a single protein with multiple functional domains or as individually-cleaved smaller proteins. RdRp, Mtr/guanylyltransferase and NTPase/RNA helicase activities have been experimentally demonstrated for ORF1 recombinant proteins.
Lipids
None reported.
Carbohydrates
Evidence for glycosylation of the major CP has been reported following its expression in mammalian cells. The CP sequence contains three potential sites for N-linked glycosylation and a signal peptide sequence at its N terminus. Mutations within the CP glycosylation sites prevent the formation of infectious virus particles, although the lethal effect is due to altered protein structure rather than elimination of glycosylation.
Genome organization and replication
The RNA genome of HEV is organized into three ORFs, with the non-structural proteins encoded toward the 5′ end of the genome and the structural protein(s) toward the 3′ end. Capped genomic RNA is infectious for chickens, pigs, rhesus monkeys and chimpanzees. The 5′-NCR is only about 26 nt long, and a cap structure has been identified in the 5′ end of the viral genome and may play a role in the initiation of hepatitis E virus replication. The 3′-NTR contains a cis-reactive element. ORF1 encodes the non-structural polyprotein. ORF2 encodes the major CP, and binds to cell surface heparan sulfate proteoglycans (HSPGs) in liver cells. The ORF3 encodes a small phosphoprotein (113–114 aa) with a multifunctional C-terminal region. ORF2 overlaps ORF3, but neither overlaps with ORF1 (Figure 4). A bicistronic subgenomic mRNA encoding both ORF2 and ORF3 proteins has been identified.
Although avian hepatitis E virus shares only about 50% nucleotide sequence identity with HEV isolates, the genomic organization and functional motifs are relatively conserved between them (Figure 4).
Antigenic properties
A single serotype of HEV has been described, with extensive cross-reactivity among circulating human and swine strains. Antibodies cross-reactive with capsid protein epitopes of human strains have been reported in various animal species but the viruses responsible for the cross-seropositivity were genetically identified only in pig, chicken, deer, mongoose, rat and rabbit. Avian hepatitis E virus also cross-reacts serologically with strains of HEV, and common antigenic epitopes have been identified in the capsid protein.
Biological properties
HEV is associated in humans with outbreaks and sporadic cases of enterically transmitted acute hepatitis. The virus is considered endemic in tropical and subtropical countries of Asia and Africa, as well as in Mexico, but antibody prevalence studies suggest a global distribution of this virus. HEV is a recognized zoonotic virus, and pigs and more likely other animal species are reservoirs.
Transmission is via the fecal–oral route. Sporadic cases of human hepatitis E have been reported in both industrialized and developing countries, although epidemics occur only in developing countries. The sources of infection appear to be different for epidemics and sporadic cases: contaminated drinking water is the main source for epidemics, whereas the risk factors for sporadic cases include shellfish, contaminated animal meats and direct contacts with infected animals. Human-to-human transmission seems rare in hepatitis E epidemics. Genotypes 1 and 2 strains are restricted to humans, whereas genotypes 3 and 4 strains have a broader host range and are zoonotic. Interspecies transmission of genotypes 3 and 4 hepatitis E virus between swine and non-human primates has been experimentally demonstrated. Pig handlers in both developing and industrialized countries are shown to be at increased risk of hepatitis E virus infection. Sporadic cases of acute hepatitis E have been epidemiologically and genetically linked to the consumption of contaminated raw and undercooked animal meats.
Avian hepatitis E virus infection in chickens is widespread and approximately 71% of chicken flocks and 30% of chickens in the United States were positive for IgG antibodies to the virus. Avian hepatitis E virus infected turkeys but failed to infect two rhesus monkeys, suggesting that the virus is likely not zoonotic. Attempts to experimentally infect mice and pigs with avian hepatitis E virus were unsuccessful. In chickens experimentally infected with avian hepatitis E virus, replicating viruses were detected in livers as well as in several extrahepatic tissues, indicating that the virus replicates not only in the liver but in the gastrointestinal tissues as well.
Genus Hepevirus
Type species Hepatitis E virus
Distinguishing features
Viruses within the genus Hepevirus infect mammals, and thus far strains within this genus have been genetically identified from human, pig, mongoose, deer, rat and rabbit.
List of species in the genus Hepevirus
| Hepatitis E virus |
|
|
| Hepatitis E virus 1 (Burma) | [M73218] | (HEV-1) |
| Hepatitis E virus 2 (Mexico) | [M74506] | (HEV-2) |
| Hepatitis E virus 3 (Meng) | [AF082843] | (HEV-3) |
| Hepatitis E virus 4 (T1) | [AJ272108] | (HEV-4) |
Species names are in italic script; names of strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Hepevirus but have not been approved as species
| Rat hepatitis E virus | [GQ504009*] | (Rat HEV) |
* Sequence does not comprise the complete genome.
A novel strain of hepevirus was detected from fecal samples of wild Norway rats (Rattus norvegicus) in Germany. Based on the available partial sequence, the rat hepevirus shares approximately 50–60% nucleotide sequence identity to human and avian strains, respectively. The rat hepevirus thus appears to be a new genotype in the genus Hepevirus.
List of unassigned species in the family Hepeviridae
| Avian hepatitis E virus |
|
|
| Avian hepatitis E virus 1 (Australia) | [AM943647] | (avian HEV-1) |
| Avian hepatitis E virus 2 (USA) | [AY535004] | (avian HEV-2) |
| Avian hepatitis E virus 3 (Europe) | [AM943646] | (avian HEV-3) |
Species names are in italic script; names of strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
Phylogenetic relationships within the family
Currently, hepatitis E virus, the only species in the genus Hepevirus, includes the four recognized major genotypes of mammalian hepeviruses: genotype 1 (Burmese-like Asian strains), genotype 2 (a single Mexican strain and some African strains), genotype 3 (strains from sporadic human cases in industrialized countries, and animal strains from pig, deer and mongoose), and genotype 4 (strains from sporadic human cases in Asia, and swine strains from pigs). A rabbit hepatitis E virus was recently identified from farmed rabbits in China. It shares 74%, 73%, 78–79% and 74–75% nucleotide sequence identity to genotypes 1, 2, 3, 4 of hepatitis E virus and 46–47% identity to avian hepatitis E virus and appears to be a distant variant of HEV genotype 3 (Figure 5A). The rat hepatitis E virus appears to be a new genotype within the genus Hepevirus (Figure 5B).
Avian hepatitis E virus is phylogenetically distinct from mammalian hepeviruses, and forms a distinct clade within the family Hepeviridae that may justify creation of a new genus. At least three genotypes of Avian hepatitis E virus have been identified from chickens worldwide (Figure 5A and 5B): genotype 1 from chickens in Australia, genotype 2 from chickens in the USA, and genotype 3 from chickens in Europe. The inter-genotype nucleotide sequence identity among the three genotypes is approximately 82%.
Similarity with other taxa
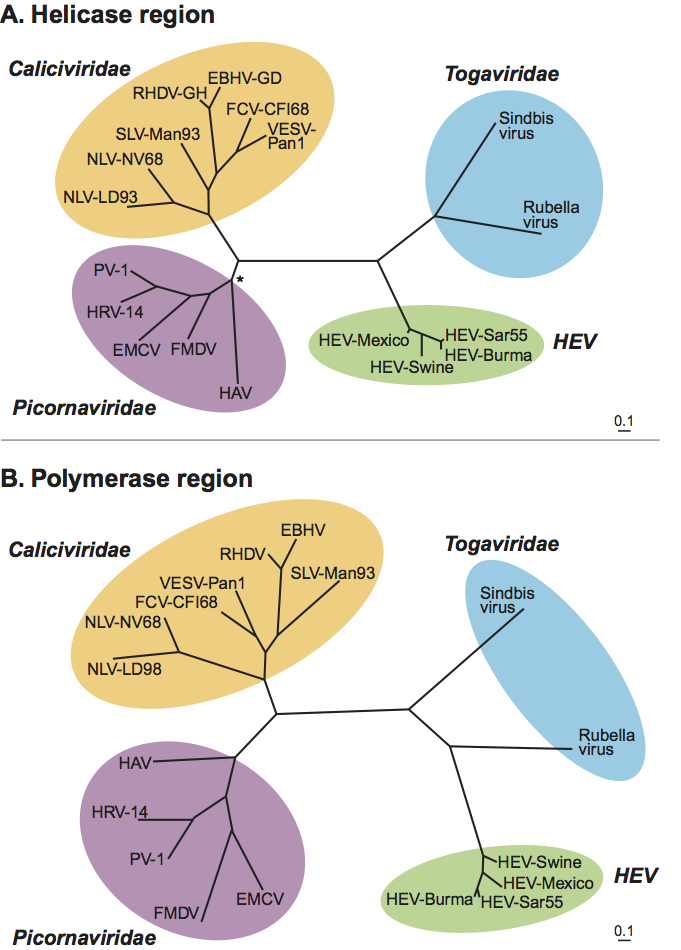
HEV is similar to members of the family Caliciviridae based on the superficial structural morphology as revealed by electron microscopy, and its genome organization. However, HEV does not share significant sequence homology with members of the family Caliciviridae. The Cap structure at the 5′ end of the HEV genome is absent from caliciviruses. HEV shows highest, but limited, amino acid sequence similarity in its replicative enzymes with Rubella virus and alphaviruses of the family Togaviridae and with plant furoviruses (Figure 6). The capping enzyme of HEV has properties very similar to those of viruses within the “alphavirus-like supergroup”.
Derivation of name
Hepe: from hepatitis E virus.
Further reading
Journals and books
Bilic et al., 2009 I. Bilic, B. Jaskulska, A. Basic, C.J. Morrow, M. Hess, Sequence analysis and comparison of avian hepatitis E viruses from Australia and Europe indicate the existence of different genotypes. J. Gen.Virol. 90 (2009) 863–873.
Emerson and Purcell, 2007 S.U. Emerson, R.H. Purcell, D.M. Knipe, P.M. Howley, Hepatitis E virusFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott Williams & Wilkins, Philadelphia, PA20073047–3058.
Haqshenas et al., 2001 G. Haqshenas, H.L. Shivaprasad, P.R. Woolcock, D.H. Read, X.J. Meng, Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J. Gen.Virol. 82 (2001) 2449–2462.
Graff et al., 2006 J. Graff, U. Torian, H. Nguyen, S.U. Emerson, A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J. Virol. 80 (2006) 5919–5926.
Johne et al., 2010 R. Johne, A. Plenge-Bönig, M. Hess, R.G. Ulrich, J. Reetz, A. Schielke, Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J. Gen.Virol. 91 (2010) 750–758.
Kalia et al., 2009 M. Kalia, V. Chandra, S.A. Rahman, D. Sehgal, S. Jameel, Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J. Virol. 83 (2009) 12714–12724.
Koonin et al., 1992 E.V. Koonin, A.E. Gorbalenya, M.A. Purdy, M.N. Rozanov, G.R. Reyes, D.W. Bradley, Computer-assisted assignment of functional domains in the non-structural polyprotein of hepatitis E virus: Delineation of an additional group of positive–strand RNA plant and animal viruses. Proc. Natl Acad. Sci., U S A. 89 (1992) 8259–8263.
Meng et al., 1997 X.J. Meng, R.H. Purcell, P.G. Halbur, J.R. Lehman, D.M. Webb, T.S. Tsareva, J.S. Haynes, B.J. Thacker, S.U. Emerson, A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl Acad. Sci., U S A. 94 (1997) 9860–9865.
Payne et al., 1999 C.J. Payne, T.M. Ellis, S.L. Plant, A.R. Gregory, G.E. Wilcox, Sequence data suggests big liver and spleen disease virus (BLSV) is genetically related to hepatitis E virus. Vet. Microbiol. 68 (1999) 119–125.
Zhao et al., 2009 C. Zhao, Z. Ma, T.J. Harrison, R. Feng, C. Zhang, Z. Qiao, J. Fan, H. Ma, M. Li, A. Song, Y. Wang, A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J. Med. Virol. 81 (2009) 1371–1379.
Websites
VIPR Virus Pathogen Resource: http://www.viprbrc.org
Contributed by
Meng, X.J., Anderson, D.A., Arankalle, V.A., Emerson, S.U., Harrison, T.J., Jameel, S. and Okamoto, H.
Figures
Figure 1 Negative contrast electron micrograph of virions of an isolate of hepatitis E virus, in the bile fluid from a monkey challenged with the genotype 2 Mexican strain of hepatitis E virus. The bar represents 100 nm.
(From Ticehurst et al. (1992). J. Infect. Dis., 165, 835845; with permission.)
Figure 2 Structure of the hepatitis E virus-like particle (VLP) (T=1). (A) Crystal structure of hepatitis E virus VLP. The three domains, S, P1 and P2 are colored blue, yellow and red, respectively. The VLP is positioned in a standard orientation with the 3 2-fold icosahedral symmetry axes aligned along the vertical, horizontal, and viewing directions, respectively. (B) Cryo-EM reconstruction at 14 resolution. The surface is colored by radial depth cue from blue, yellow, to red. (C) Hepatitis E virus VLP with only the S domain. (D) VLP with S and P1 domains. (E) VLP with P1 and P2 domains.
(From Guu et al. (2009). Proc. Natl Acad. Sci., U S A, 106, 1299212997; with permission.)
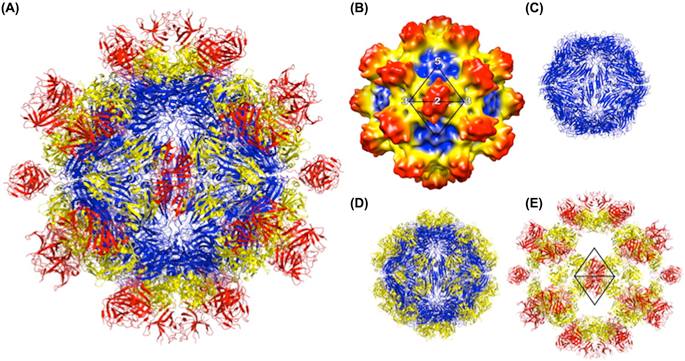
Figure 3 Electron micrograph of 3035 nm diameter particles of Avian hepatitis E virus. The virus particles were detected from a bile sample of a chicken with hepatitissplenomegaly syndrome. Bar=100 nm.
(From Haqshenas et al. (2001). J. Gen. Virol., 82, 24492462; with permission.)
Figure 4 Schematic diagram of the genomic organization of hepatitis E virus: a short 5 non-coding region (NCR), a 3 NCR, and three ORFs. ORF2 and ORF3 overlap each other but neither overlaps ORF1. ORF1 encodes non-structural proteins including putative functional domains; ORF2 encodes capsid protein and ORF3 encodes a small phosphoprotein with a multi-functional C-terminal region. MT, methytransferase; Y, Y domain; P, a papain-like cystein protease; HVR, a hypervariable region that is dispensable for virus infectivity; Hel, helicase; RdRp, RNA-dependent RNA polymerase
(From Meng, X.J. (2008). Hepatitis E virus (Hepevirus). In: Encyclopedia of Virology (5 vols), 3rd edn (B.W.J. Mahy and M.H.V. van Regenmortel, Eds.), Oxford, Elsevier, pp. 377383; with permission.)
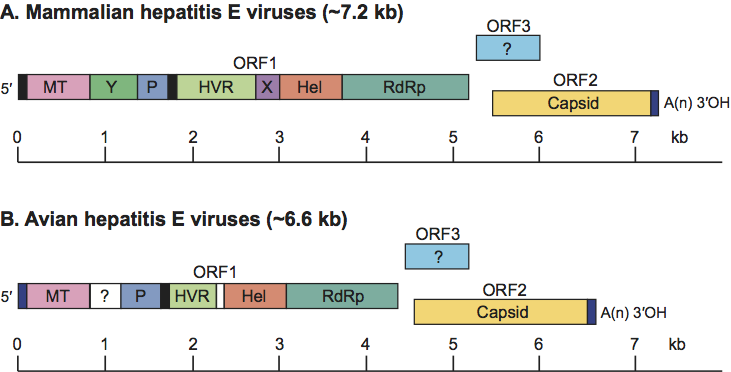
Figure 5 Phylogenetic trees depicting the relationship between strains of mammalian hepatitis E virus in the genus Hepevirus and the unassigned species Avian hepatitis E virus (courtesy of Hiroaki Okamoto). (A) A phylogenetic tree based on the full-length genomic sequences of representative hepatitis E virus strains including the four major genotypes of mammalian hepatitis E virus, the newly-identified rabbit hepatitis E virus and the three genotypes of Avian hepatitis E virus; (B) A phylogenetic tree based upon partial sequence (1545 nt) of the rat hepatitis E virus along with other hepatitis E virus strains in Figure 6A. GenBank accession numbers for the strains used in these analyses are Burma hHEV (M73218); Morocco hHEV (AY230202); Mexico hHEV (M74506); USA hHEV (AF060669); USA sHEV (AF082843); Japan gt3-hHEV1 (AP003430); Japan gt3-sHEV (AB073912); Japan gt3-hHEV2 (AB248520); Kyrgyzstan sHEV (AF455784); China hHEV (AJ272108); Japan gt4-sHEV (AB097811); Japan gt4-hHEV (AB220973); USAaHEV (AY535004); aavUSAaHEV (EF206691); AaHEV (AM943647); EaHEV (AM943646); rabbit HEV1 (FJ906895); rabbit HEV2 (FJ906896); rat HEV1 (GQ504009); and rat HEV2 (GQ504010).
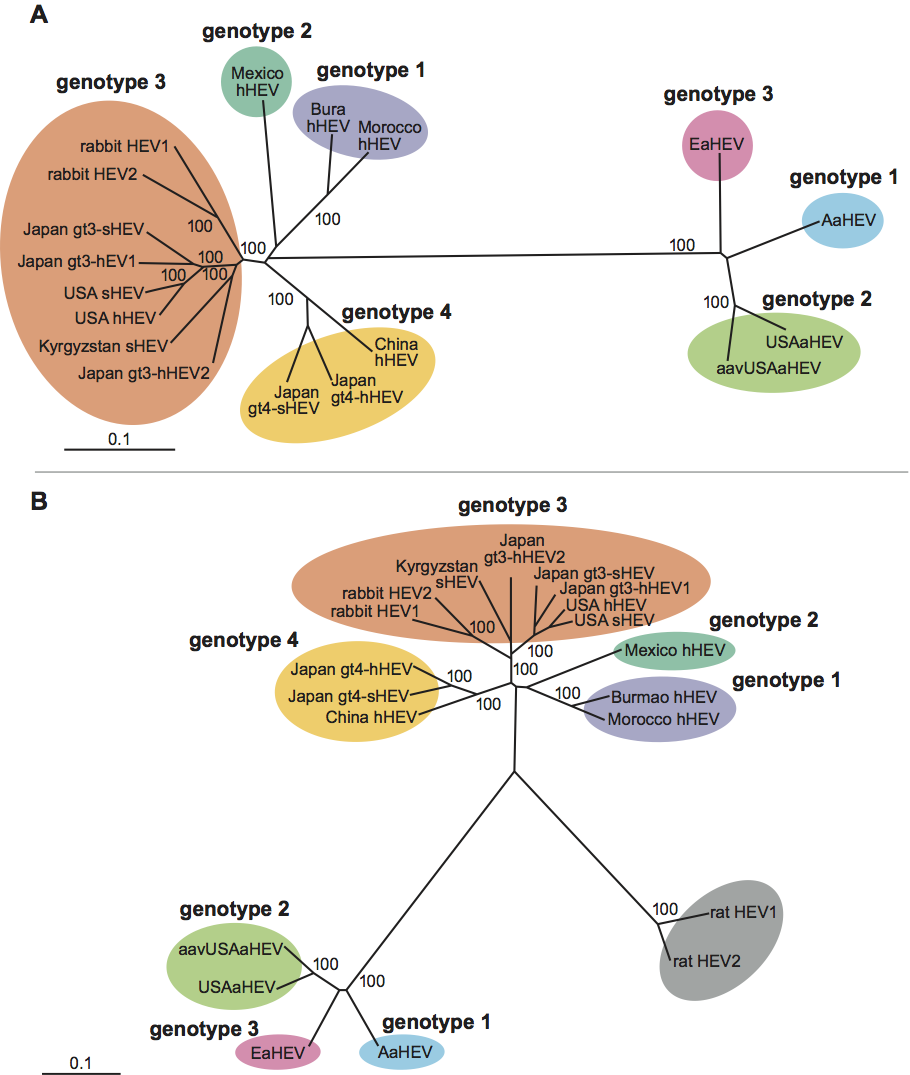
Figure 6 Phylogenetic relationships of hepatitis E virus with members of the families Picornaviridae, Caliciviridae and Togaviridae. The helicase (Hel) and polymerase (Pol) regions of the genome were analyzed (courtesy of T. Berke and D.O. Matson). (A) Partial gene sequences (200 aa) from the proposed helicase region were used for the phylogenetic analysis and included representative strains from each family. Clustal W v1.7 was used to create a multiple alignment for the aa sequences, which was verified by alignment of known motifs in the region (e.g. GxGKS/T). The nt sequences were added and aligned by hand using the corresponding aa sequences as a template resulting in a consensus length of 608 nt. A phylogenetic tree was constructed from the nt sequence alignment using the maximum likelihood algorithm in the program DNAML from the PHYLIP 3.52c package within UNIX environment. For the algorithm, the global rearrangement option was invoked and the order of sequence input was randomized ten times. Other menu options were left as default. The resultant tree is unrooted and the phylogenetic distances are in the unit of expected number of substitutions per site. Branch points of the resulting tree had a confidence level of P<0.01 (P<0.05*). GenBank accession numbers for the strains in this analysis were M87661 (Norwalk virus, NLV-NV68), X86557 (Lordsdale virus, NLV-LD93), U52086 (primate calicivirus, VESV-Pan1), U13992 (feline calicivirus, FCV-CFI/68), Z69620 (European brown hare syndrome virus, EBHV-GD), M67473 (rabbit hemorrhagic disease virus, RHDV-GH), X86560 (Sapporo virus, SLV-Man93), J02281 (human poliovirus 1, PV-1), K02121 (Human rhinovirus type 14, HRV-14), M22458 (encephalomyocarditis virus, EMCV), X00429 (foot-and-mouth disease virus, FMDV), K02990 (hepatitis A virus, HAV), M15240 (rubella virus), J02363 (Sindbis virus), M73218 (hepatitis E virus, HEV-Burma), M80581 (hepatitis E virus, HEV-Sar55), M74506 (hepatitis E virus, HEV-Mexico), AF011921 (hepatitis E virus, HEV-Swine). (B) Partial gene sequences (200 aa) from the proposed polymerase region were used for the phylogenetic analysis and included representative strains from each family. Clustal W v1.7 was used to create a multiple alignment for the aa sequences, which was verified by alignment of known motifs in the region (e.g. SGxxxTxxxMT/S, GDD). The nt sequences were added and aligned by hand using the corresponding aa sequences as template resulting in a consensus length of 590 nt. A phylogenetic tree was constructed as described above and GenBank accession numbers for the strains in this analysis were identical to those above.