Updated ICTV Online (10th) Report Chapter Available
Follow this link to the current Flaviviridae Online Report
Family: Flaviviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are 40–60 nm in diameter, spherical in shape and contain a lipid envelope. The capsid is composed of a single protein and the envelope contains two or three virus-encoded membrane proteins. Specific descriptions of the three individual genera and a tentative fourth genus are given in the corresponding sections.
Physicochemical and physical properties
The virion Mr, buoyant density, sedimentation coefficient and other physicochemical properties differ among the members of the genera and are described separately in the corresponding sections.
Nucleic acid
Genomes are positive sense ssRNA of approximately 11, 12.3 and 9.6 kb for members of the genera Flavivirus, Pestivirus and Hepacivirus, respectively. All members of the family lack a 3′-terminal poly(A) tract. Only the genomes of flaviviruses contain a 5′-terminal type I cap structure.
Proteins
The virions of members of the family have a single, small basic capsid (C) and two (flavivirus and hepacivirus) or three (pestivirus) membrane-associated envelope proteins. Viruses that are candidates for inclusion in a possible fourth genus appear to lack a complete nucleocapsid protein gene. The nonstructural proteins contain sequence motifs characteristic of a serine protease, RNA helicase and RdRp that are encoded at similar locations along the genome in all genera. Further details of specific functional properties are given in the corresponding sections of the individual genera.
Lipids
Lipids present in virions are derived from host cell membranes and make up 17% of the total virion weight in the case of flaviviruses. The lipid content of pesti- and hepaciviruses has not been determined.
Carbohydrates
Virions contain carbohydrates in the form of glycolipids and glycoproteins.
Genome organization and replication
The genomic RNA of all members of the family has a similar organization and is the viral mRNA found in infected cells. It contains a single long ORF flanked by 5′- and 3′-terminal non-coding regions (NCRs) that form specific secondary structures required for genome replication and translation. Flaviviruses, but not pestiviruses or hepaciviruses, produce a unique, subgenomic, small (0.3–0.5 kb) non-coding RNA derived from 3′-NCR of genomic RNA, which is essential for virus replication in cells and modulates pathogenicity in animals. Translation-initiation of genomic RNA is cap-dependent in the case of flaviviruses, whereas IRES elements are present in the other genera. Viral proteins are synthesized as part of a polyprotein that is co- and post-translationally cleaved by viral and cellular proteases. The structural proteins are contained in the N-terminal portion of this polyprotein and the non-structural proteins in the remainder. The latter include a serine protease, an RNA helicase and the RdRp. RNA synthesis occurs in the cytoplasm in association with modified cellular membranes via synthesis of full-length negative-strand intermediates. Virion assembly, including acquisition of a glycoprotein-containing lipid envelope, occurs by budding through intracellular membranes. Viral particles are transported in cytoplasmic vesicles through the secretory pathway before they are released by exocytosis, as shown for members of the genus Flavivirus and assumed for members of the other genera.
Antigenic properties
The genera are antigenically unrelated, but serological cross-reactivity exists among members within each genus.
Biological properties
The biological properties of the three genera exhibit different characteristics and are described in the corresponding sections.
Genus Flavivirus
Type species Yellow fever virus
Distinguishing features
The 5′ end of the genome possesses a type I cap (m7GpppAmp) not seen in the other genera. Most flaviviruses are transmitted to vertebrate hosts by arthropod vectors, mosquitoes or ticks, in which they actively replicate. Some flaviviruses are zoonotic agents transmitted between rodents or bats without known arthropod vectors.
Virion properties
Morphology
Virions are 50 nm in diameter and spherical in shape (Figure 1). Two virus forms can be distinguished. Mature virions contain two virus encoded membrane-associated proteins, E and M. Intracellular immature virions contain the precursor prM instead of M, which is proteolytically cleaved during maturation. In certain instances, partially mature/immature forms are also released from infected cells. The atomic structure of the major envelope protein E from tick-borne encephalitis virus (TBEV), dengue virus (DENV) and West Nile virus (WNV) has been determined by X-ray crystallography. It is a dimeric, rod-shaped molecule that is oriented parallel to the membrane and does not form spike-like projections in its neutral pH conformation. Image reconstructions from cryo-electron micrographs (Figure 1) have shown that the virion envelope has icosahedral symmetry, in which E protein dimers are organized in a herringbone-like arrangement.
Physicochemical and physical properties
Virion Mr has not been precisely determined. Mature virions sediment at about 200S and have a buoyant density of about 1.19 g cm−3 in sucrose. Viruses are stable at slightly alkaline pH 8.0 but are readily inactivated at acidic pH, temperatures above 40 °C, organic solvents, detergents, ultraviolet light and gamma-irradiation.
Nucleic acid
The virion RNA of flaviviruses is a positive sense infectious ssRNA of about 11 kb. The 5′ end of the genome possesses a type I cap (m-7GpppAmp) followed by the conserved dinucleotide AG. The 3′ ends lack a terminal poly(A) tract and terminate with the conserved dinucleotide CU.
Proteins
Virions contain three structural proteins: C (11 kDa), E (50 kDa), the major envelope protein, and either prM (26 kDa), in immature virions, or M (8 kDa), in mature virions. The E protein is the viral hemagglutinin, which mediates both receptor binding and acid pH-dependent fusion activity after uptake by receptor-mediated endocytosis. Seven nonstructural proteins are synthesized in infected cells: NS1 (46 kDa), NS2A (22 kDa), NS2B (14 kDa), NS3 (70 kDa), NS4A (16 kDa), NS4B (27 kDa) and NS5 (103 kDa). NS1 has multiple forms and roles, with a cell-associated form functioning in viral RNA replication and a secreted form that regulates complement activation. One such form, a NS1′ protein, is the product of a −1 ribosomal frameshift and plays a role in viral neuroinvasiveness. NS3 is a multifunctional protein. The N-terminal one-third of the protein forms the viral serine protease complex together with NS2B that is involved in processing the polyprotein. The C-terminal portion of NS3 contains an RNA helicase domain involved in RNA replication, as well as an RNA triphosphatase activity that is probably involved in formation of the 5′-terminal cap structure of the viral RNA. NS5 is the largest and most highly conserved flavivirus protein. It is a multifunctional protein that acts as the viral RdRp and also possesses methyltransferase activity involved in the modification of the viral cap structure.
Lipids
Virions contain about 17% lipid by weight; lipids are derived from host cell membranes.
Carbohydrates
Virions contain about 9% carbohydrate by weight (glycolipids, glycoproteins); their composition and structure are dependent on the host cell (vertebrate or arthropod). N-glycosylation sites are present in the proteins prM (1 to 3 sites), E (0 to 2 sites) and NS1 (1 to 3 sites).
Genome organization and replication
The genomic RNA represents the only viral messenger RNA in flavivirus-infected cells. It consists of a single long ORF of more than 10,000 nt that codes for all structural and non-structural proteins and is flanked by NCRs at the 5′- and 3′-terminal ends (Figure 2).
Both the 5′-NCR and the 3′-NCR contain RNA sequence motifs that are involved in viral RNA translation, replication and possibly packaging. Although RNA secondary structure and function of some elements are conserved, sequence composition, length and exact localization can vary considerably between different members of the genus, in particular between tick-borne and mosquito-borne flaviviruses. In some cases, the 3′-NCR of tick-borne encephalitis virus contains an internal poly(A) tract. Flavivirus infection induces dramatic rearrangements of cellular membrane structures within the perinuclear endoplasmic reticulum (ER) and causes the formation of ER-derived vesicular packets that most likely represent the sites of viral replication. After translation of the incoming genomic RNA, RNA replication begins with synthesis of complementary negative strands, which are then used as templates to produce additional genome-length positive-stranded molecules. These are synthesized by a semi-conservative mechanism involving replicative intermediates (containing double stranded regions as well as nascent single stranded molecules) and replicative forms (duplex RNA molecules). Translation usually starts at the first AUG of the ORF, but may also occur at a second in-frame AUG located 12 to 14 codons downstream in mosquito-borne flaviviruses. The polyprotein is processed by cellular proteases and the viral NS2B-NS3 serine protease to give rise to the mature structural and nonstructural proteins. Protein topology with respect to the ER and cytoplasm is determined by internal signal and stop-transfer sequences. Virus particles can first be observed in the rough endoplasmic reticulum, which is believed to be the site of virus assembly. These immature virions are then transported through the membrane systems of the host secretory pathway to the cell surface where exocytosis occurs. Shortly before virion release, the prM protein is cleaved by furin or a furin-like cellular protease to generate mature virions. Flavivirus-infected cells also release a noninfectious subviral particle that has a lower sedimentation coefficient than whole virus (70S vs. 200S) and exhibits hemagglutination activity (slowly sedimenting hemagglutinin; SHA).
Antigenic properties
All flaviviruses are serologically related, which can be demonstrated by binding assays such as ELISA and by hemagglutination-inhibition using polyclonal and monoclonal antibodies. Neutralization assays are more discriminating and have been used to define several serocomplexes of more closely related flaviviruses (see “List of species in the genus”). The envelope protein E is the major target for neutralizing antibodies and induces protective immunity. The E protein also induces flavivirus cross-reactive non-neutralizing antibodies. Antigenic sites involved in neutralization have been mapped to each of the three structural domains of the E protein. Antibodies to prM can also mediate immunity, probably by neutralizing viruses with partially uncleaved prM. The NS1 protein can also induce antibodies that protect infected animals from lethal infection.
Biological properties
Host range
Flaviviruses can infect a variety of vertebrate species and in many cases arthropods. Some viruses have a limited vertebrate host range (e.g., only primates), others can infect and replicate in a wide variety of species (mammals, birds, etc.). Arthropods are usually infected when they feed on a vertebrate host during viremia, but non-viremic transmission has also been described for tick-borne flaviviruses. A new group of flaviviruses that appear only to infect mosquitoes is now recognized. The prototype of these flaviviruses was tentatively assigned to the genus Flavivirus following the discovery of cell fusing agent virus. However, several genetically related but distinct insect-only flaviviruses have now been identified and will need to be considered as a possible separate group of viruses within the genus.
Transmission
Most flaviviruses are arthropod-borne viruses that are maintained in nature by transmission from hematophagous arthropod vectors to vertebrate hosts. About 50% of known flaviviruses are mosquito-borne, 28% are tick-borne and the rest are zoonotic agents transmitted between rodents or bats without known arthropod vectors. For some flaviviruses, the transmission cycle has not yet been identified. In certain instances, flaviviruses can be transmitted to humans by blood products, organ transplantation, non-pasteurized milk or aerosols. In the arthropod vectors, the viruses may also be passed on trans-ovarially or vertically (mosquitoes, ticks) and trans-stadially (ticks). The mechanisms of virus transmission involving the insect-only flaviviruses may include vertical transmission but other mechanisms need to be considered to explain the success with which these viruses appear to have dispersed globally.
Geographical distribution
Flaviviruses have a world-wide distribution but individual species are restricted to specific endemic or epidemic areas (e.g., yellow fever virus in tropical and subtropical regions of Africa and South America; dengue virus in tropical areas of Asia, Oceania, Africa, Australia and the Americas; Japanese encephalitis virus in South-East Asia; tick-borne encephalitis virus in Europe and Northern Asia).
Pathogenicity
More than 50% of known flaviviruses have been associated with human disease, including the most important human pathogens: yellow fever virus, dengue virus, Japanese encephalitis virus, West Nile virus and tick-borne encephalitis virus. Flavivirus-induced diseases may be associated with symptoms of the central nervous system (e.g., meningitis, encephalitis), fever, arthralgia, rash and hemorrhagic fever. Several flaviviruses are pathogenic for domestic or wild animals (turkey, pig, horse, sheep, dog, grouse, muskrat) and cause economically important diseases.
Species demarcation criteria in the genus
Species demarcation criteria in the genus include:
- Nucleotide and deduced amino acid sequence data.
- Antigenic characteristics.
- Geographic association.
- Vector association.
- Host association.
- Disease association.
- Ecological characteristics.
Species demarcation considers a combination of each of the criteria listed above. While nucleotide sequence relatedness and the resulting phylogenies are important criteria for species demarcation, the other listed criteria may be particularly useful in demarcation of genetically closely related viruses. For example far-eastern (FE) strains of tick-borne encephalitis virus exhibit distinct ecological differences when compared with Omsk hemorrhagic fever virus (OHFV) despite the fact that they are genetically relatively closely related. TBEV-FEs are associated predominantly with Ixodes persulcatus ticks in forest environments in far-east Russia, whereas OHFV is found in the Steppe regions of western Siberia associated particularly with Dermacentor spp. and to a lesser extent with Ixodes spp. These viruses are also antigenically distinguishable in neutralization tests that employ convalescent sera.
Louping ill virus (LIV) and TBEV provide another example where, despite their close genetic relationships and similar host ranges, they display different ecologies (moorlands versus forests), pathogenicities (red grouse, sheep/goats versus humans) and geographical distributions (UK versus Europe/Eurasia), thus justifying their classification as distinct species.
On the other hand (like poliovirus with its three serotypes), the four dengue virus serotypes comprise a single species, despite being phylogenetically and antigenically quite distinct. This is justified by the fact that they co-circulate in the same geographical areas and ecological habitats, and that they exploit identical vectors, exhibit similar life cycles and disease manifestations.
List of species in the genus Flavivirus
| Tick-borne | ||
| Mammalian tick-borne virus group: |
|
|
| Gadgets Gully virus |
|
|
| Gadgets Gully virus | [DQ235145] | (GGYV) |
| Kyasanur Forest disease virus |
|
|
| Kyasanur Forest disease virus | [AY323490] | (KFDV) |
| Alkhurma hemorrhagic fever virus | [AF331718] | (AHFV) |
| Langat virus |
|
|
| Langat virus | [AF253419] | (LGTV) |
| Louping ill virus |
|
|
| British subtype | [D12937] | (LIV-Brit) |
| Irish subtype | [X86784] | (LIV-Ir) |
| Spanish subtype | [DQ235152 | (LIV-Spain) |
| Turkish sheep encephalitis virus subtype | [DQ235151] | (TSEV) |
| Greek goat encephalitis virus subtype | [DQ235153] | (GGEV) |
| Omsk hemorrhagic fever virus |
|
|
| Omsk hemorrhagic fever virus | [AY323489] | (OHFV) |
| Powassan virus |
|
|
| Powassan virus | [L06436] | (POWV) |
| Royal Farm virus |
|
|
| Royal Farm virus | [DQ235149] | (RFV) |
| Tick-borne encephalitis virus |
|
|
| European subtype | [M27157, M33668] | (TBEV-Eur) |
| Far Eastern subtype | [X07755] | (TBEV-FE) |
| Siberian subtype | [L40361] | (TBEV-Sib) |
| Seabird tick-borne virus group: |
|
|
| Meaban virus |
|
|
| Meaban virus | [DQ235144] | (MEAV) |
| Saumarez Reef virus |
|
|
| Saumarez Reef virus | [DQ235150] | (SREV) |
| Tyuleniy virus |
|
|
| Tyuleniy virus | [DQ235148] | (TYUV) |
| Kadam virus group (probably tick-borne): |
|
|
| Kadam virus |
|
|
| Kadam virus | [DQ235146] | (KADV) |
| Mosquito-borne | ||
| Aroa virus group: |
|
|
| Aroa virus |
|
|
| Aroa virus | [AF013362] | (AROAV) |
| Bussuquara virus | [AF013366] | (BSQV) |
| Iguape virus | [AF013375] | (IGUV) |
| Naranjal virus | [AF013390] | (NJLV) |
| Dengue virus group: |
|
|
| Dengue virus |
|
|
| Dengue virus 1 | [U88536] | (DENV-1) |
| Dengue virus 2 | [M19197] | (DENV-2) |
| Dengue virus 3 | [M93130] | (DENV-3) |
| Dengue virus 4 | [AF326573] | (DENV-4) |
| Japanese encephalitis virus group: |
|
|
| Cacipacore virus |
|
|
| Cacipacore virus | [AF013367] | (CPCV) |
| Japanese encephalitis virus |
|
|
| Japanese encephalitis virus | [M18370] | (JEV) |
| Koutango virus |
|
|
| Koutango virus | [AF013384] | (KOUV) |
| Murray Valley encephalitis virus |
|
|
| Alfuy virus | [AF013360] | (ALFV) |
| Murray Valley encephalitis virus | [AF151266] | (MVEV) |
| St Louis encephalitis virus |
|
|
| St. Louis encephalitis virus | [DQ525916] | (SLEV) |
| Usutu virus |
|
|
| Usutu virus | [AF013412] | (USUV) |
| West Nile virus |
|
|
| Kunjin virus | [D00246] | (KUNV) |
| West Nile virus | [M12294] | (WNV) |
| Yaounde virus |
|
|
| Yaounde virus | [AF013413] | (YAOV) |
| Kokobera virus group: |
|
|
| Kokobera virus |
|
|
| Kokobera virus | [AF013383] | (KOKV) |
| Stratford virus | [AF013407] | (STRV) |
| Ntaya virus group: |
|
|
| Bagaza virus |
|
|
| Bagaza virus | [AF013363] | (BAGV) |
| Ilheus virus |
|
|
| Ilheus virus | [AF013376] | (ILHV) |
| Rocio virus | [AF013397] | (ROCV) |
| Israel turkey meningoencephalitis virus |
|
|
| Israel turkey meningoencephalitis virus | [AF013377] | (ITV) |
| Ntaya virus |
|
|
| Ntaya virus | [AF013392] | (NTAV) |
| Tembusu virus |
|
|
| Tembusu virus | [AF013408] | (TMUV) |
| Zika virus |
|
|
| Zika virus | [DQ859059] | (ZIKV) |
| Yellow fever virus group: |
|
|
| Sepik virus |
|
|
| Sepik virus | [DQ859063] | (SEPV) |
| Wesselsbron virus |
|
|
| Wesselsbron virus | [DQ859058] | (WSLV) |
| Yellow fever virus |
|
|
| Yellow fever virus | [X03700] | (YFV) |
| Probably mosquito-borne | ||
| Kedougou virus group: |
|
|
| Kedougou virus |
|
|
| Kedougou virus | [DQ859061] | (KEDV) |
| Edge Hill virus group: |
|
|
| Banzi virus |
|
|
| Banzi virus | [DQ859056] | (BANV) |
| Bouboui virus |
|
|
| Bouboui virus | [DQ859057] | (BOUV) |
| Edge Hill virus |
|
|
| Edge Hill virus | [DQ859060] | (EHV) |
| Jugra virus |
|
|
| Jugra virus | [DQ859066] | (JUGV) |
| Saboya virus |
|
|
| Potiskum virus | [DQ859067] | (POTV) |
| Saboya virus | [DQ859062] | (SABV) |
| Uganda S virus |
|
|
| Uganda S virus | [DQ859065] | (UGSV) |
| Species with no known arthropod vector | ||
| Entebbe bat virus group: |
|
|
| Entebbe bat virus |
|
|
| Entebbe bat virus | [AF013373] | (ENTV) |
| Sokoluk virus | [AF013405] | (SOKV) |
| Yokose virus |
|
|
| Yokose virus | [AF013414] | (YOKV) |
| Modoc virus group: |
|
|
| Apoi virus |
|
|
| Apoi virus | [AF160193] | (APOIV) |
| Cowbone Ridge virus |
|
|
| Cowbone Ridge virus | [AF013370] | (CRV) |
| Jutiapa virus |
|
|
| Jutiapa virus | [AF013379] | (JUTV) |
| Modoc virus |
|
|
| Modoc virus | [AJ242984] | (MODV) |
| Sal Vieja virus |
|
|
| Sal Vieja virus | [AF013401] | (SVV) |
| San Perlita virus |
|
|
| San Perlita virus | [AF013402] | (SPV) |
| Rio Bravo virus group: |
|
|
| Bukalasa bat virus |
|
|
| Bukalasa bat virus | [AF013365] | (BBV) |
| Carey Island virus |
|
|
| Carey Island virus | [AF013368] | (CIV) |
| Dakar bat virus |
|
|
| Dakar bat virus | [AF013371] | (DBV) |
| Montana myotis leukoencephalitis virus |
|
|
| Montana myotis leukoencephalitis virus | [AJ299445] | (MMLV) |
| Phnom Penh bat virus |
|
|
| Batu Cave virus | [AF013369] | (BCV) |
| Phnom Penh bat virus | [AF013394] | (PPBV) |
| Rio Bravo virus |
|
|
| Rio Bravo virus | [AF144692] | (RBV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses that may be members of the genus Flavivirus but have not been approved as species
| Mammalian tick-borne | ||
| Karshi virus | [DQ235147] | (KSIV) |
| Mosquito-borne | ||
| Spondweni virus | [DQ859064] | (SPOV) |
| Probably arthropod-borne | ||
| Aedes flavivirus | [AB488408] | (AEFV) |
| Cell fusing agent virus | [M91671] | (CFAV) |
| Culex flavivirus | [GQ165808] | (CXFV) |
| Kamiti River virus | [AY149905] | (KRV) |
| Nakiwogo virus | [GQ165809] | (NAKV) |
| Quang Binh virus | [FJ644291] | (QBV) |
| Viruses with no known arthropod vector | ||
| Chaoyang virus | [FJ883471] | (CHAOV) |
| Lammi virus | [FJ606789] | (LAMV) |
| Ngoye virus | [DQ400858] | (NGOV) |
| Nounané virus | [EU159426] | (NOUV) |
| Tamana bat virus | [AF286080] | (TABV) |
Genus Pestivirus
Type species Bovine viral diarrhea virus 1
Distinguishing features
Relative to the other genera, pestiviruses encode two unique gene products, namely Npro and Erns. The first protein of the ORF, nonstructural protein Npro, which possesses an autoproteolytic activity and is responsible for its release from the nascent polyprotein, is not essential for virus replication in cell culture. One of the three viral envelope glycoproteins, Erns, possesses an intrinsic RNase activity. Both of these unique pestivirus proteins are involved in repression of the host type I IFN response. Two biotypes of pestiviruses, cytopathogenic (cp) and non-cytopathogenic (noncp) viruses, are distinguished by their ability to cause cytopathic effects in cell culture.
Virion properties
Morphology
Virions are 40–60 nm in diameter and spherical in shape (Figure 3). The virion envelope has 10–12 nm ring-like subunits on its surface. Structure and symmetry of the core have not been characterized.
Physicochemical and physical properties
Virion Mr has not been determined precisely. Buoyant density in sucrose is 1.10–1.15 g cm−3; S20,W is 140–150S. Virion infectivity is stable over a relatively broad pH range, but unstable at temperatures above 40 °C. Organic solvents and detergents rapidly inactivate these viruses.
Nucleic acid
The virion RNA is a positive sense, infectious molecule of ssRNA about 12.3 kb in size. The 5′-NCR contains an IRES and is about 370–385 nt in length. The 3′-NCR, with about 185–273 nt, is complex and contains a region with variable sequences and a highly conserved terminal region. Genomic RNA contains a single ORF spanning the viral genome. For some cp pestivirus strains, a small and variable segment of host cell or viral nucleic acid is integrated into particular regions (often within NS2 or directly upstream of NS3) of the viral genome, sometimes accompanied by viral gene duplications or deletions. Other cp pestiviruses contain only viral gene duplications involving all or part of the Npro and NS3 to NS4B protein-coding regions, resulting in genomic RNA of up to about 16.5 kb. In all cases, the single large ORF is maintained. Finally, cp viruses may also arise by deletion of large portions of their genomes. Such defective genomes can be rescued by co-infecting intact helper viruses.
Proteins
Virions are composed of four structural proteins: a basic nucleocapsid core protein, C (14 kDa) and three envelope glycoproteins, Erns (gp44/48), E1 (gp33) and E2 (gp55). All three glycoproteins exist as intermolecular disulfide-linked complexes: Erns homodimers, E1-E2 heterodimers and E2 homodimers. The Erns protein possesses an intrinsic RNase activity. Pestiviruses encode eight nonstructural (NS) proteins among which Npro (23 kDa), p7 (7 kDa) and NS2 (40 kDa) are not necessary for RNA replication. Npro is a proteinase that auto-catalytically releases itself from the nascent polyprotein. Nonstructural protein p7 is presumed to have a role in virus maturation. NS2-3 (120 kDa) is a multifunctional protein. The N-terminal 40% (NS2) is hydrophobic and contains a zinc finger motif for binding of a divalent metal ion. NS2 is a cysteine protease that is responsible for processing of NS2-3 to give rise to NS2 and NS3. NS3 (80 kDa) acts as both a serine protease involved in polyprotein processing and an RNA helicase/NTPase involved in RNA replication. NS2-3 is found after infection with all pestiviruses. In cells infected with cp pestiviruses, large amounts of NS3 can be detected. For noncp BVDV, noncp BDV and CSFV strains, only a minor fraction of NS2-3 is cleaved into NS2 and NS3, so that sometimes the cleavage products are difficult to detect. The NS4A (7 kDa) protein acts as a cofactor to the NS3 protease activity. The role of NS4B (33 kDa) is unknown. NS5A (58 kDa) represents a phosphorylated protein and presumably also plays a yet to be identified role in RNA replication. NS5B (75 kDa) possesses RdRp activity.
Lipids
The viruses are enveloped, but no reports have described the lipid composition.
Carbohydrates
All virus envelope glycoproteins contain N-linked glycans.
Genome organization and replication
The genomic RNA contains a single large ORF encoding a polyprotein of about 3900 aa that is preceded by a 5′-NCR of about 370–385 nt and followed by a 3′-NCR of about 185–273 nt. The gene order is 5′-Npro-C-Erns-E1-E2-p7-NS2-3(NS2-NS3)-NS4A-NS4B-NS5A-NS5B-3′ (Figure 4).
Pestivirus replication is initiated by receptor-mediated endocytosis involving most likely more than one cell surface molecule and viral glycoproteins Erns and E2. CD46 has been shown to function as a cellular receptor for BVDV but is not by itself sufficient to mediate infection. After endocytosis and uncoating, the genome RNA serves as mRNA; there are no subgenomic mRNA molecules. Translation initiation occurs by a cap-independent internal initiation mechanism involving an IRES within the 5′-NCR of the RNA. Polyprotein processing occurs co- and post-translationally by both cellular and viral proteases. Nonstructural protein Npro, the first protein of the ORF, auto-proteolytically removes itself from the nascent polyprotein by cleavage at the Npro/C site. Downstream cleavages that produce structural proteins C, Erns, E1 and E2 as well as p7 are mediated by cellular signal peptide peptidase and signal peptidase(s). Glycoprotein translocation to the endoplasmic reticulum occurs by an internal signal sequence, within the C-terminal region of the C protein. Cleavage between E2 and p7 is not complete, leading to two intracellular forms of E2 with different C-termini. Depending on the pestivirus biotype, NS2-3 either remains mostly intact or is found at reduced levels together with high amounts of its N- and C-terminal products NS2 and NS3. The increased generation of NS3 in cp pestiviruses is in most cases due to RNA recombination. Most cp pestiviruses have gene insertions, deletions, duplications or rearrangements that result in enhanced NS3 production. The NS3/NS2-3 serine protease activity is responsible for all processing events downstream of NS3. NS4A facilitates cleavages by the NS3 protease of sites 4B/5A and 5A/5B.
RNA replication occurs most likely in association with intracytoplasmic membranes, presumably in a replication complex composed of viral RNA and viral nonstructural proteins. Nonstructural proteins NS3, 4A, 4B, 5A and 5B are necessary for RNA replication; only NS5A can be provided in trans. Replicative forms of pestiviral RNA have been detected. The ratio of positive-to-negative sense RNA in cells 12 hours post-infection is about 10. RNA synthesis is resistant to actinomycin D. Virus maturation is poorly understood. However, viral proteins are not found on the cell surface, suggesting that viruses mature in intracellular vesicles and are released by exocytosis. Considerable amounts of infectious virus remain cell-associated. Host cell RNA and protein synthesis continues throughout infection.
Antigenic properties
Pestiviruses are antigenically related, and cross-reactive epitopes are found for all members. Separate antigenic determinants defined by monoclonal antibodies (Mabs) have also been identified. Antigenic variation is particularly pronounced among isolates of BVDV and BDV. The N-terminal portion of E2 contains an antigenically hypervariable region. Mab binding patterns are generally consistent with the genetic relatedness of viruses.
Infected animals mount potent antibody responses to two structural glycoproteins (Erns, E2) and to the NS2-3/NS3 protein, while antibody responses to other virus-encoded polypeptides are weak or not detectable. Erns and E2 are able to induce protection independently. Mabs reactive with these proteins can neutralize virus infectivity.
Biological properties
Pestiviruses infect pigs and ruminants, including cattle, sheep, goats and wild ruminants. There are no invertebrate hosts. Transmission occurs by direct and indirect contact (e.g., nasal or urine secretion, ***, contaminated food, etc.). Transplacental transmission occurs in all host species. Pestivirus infections may be subclinical or produce a range of clinical conditions including acute diarrhea, acute hemorrhagic syndrome, acute fatal disease, and a wasting disease. Transplacental infection can result in fetal death, congenital abnormalities, or lifelong persistent infection. Fatal mucosal disease can occur in cattle persistently infected with noncp viruses when a cp virus is generated by mutation or introduced by superinfection. Pestivirus infections of livestock are economically important worldwide.
Experimental infection models have not been established for bovine or ovine pestiviruses outside their natural mammalian hosts. However, CSFV can be adapted to propagate in rabbits. Cells derived from natural host species (bovine, porcine, ovine) support virus replication. Most virus isolates are noncp and can establish persistent infections in cell culture. Infectious noncp BVDV is often present in bovine serum products used for cell culture. Cp pestiviruses induce extensive cytopathology and form virus plaques under appropriate conditions. Death of cp pestivirus infected cells is due to apoptosis. No hemagglutinating activity has been found associated with pestiviruses.
Species demarcation criteria in the genus
Species demarcation criteria in the genus include:
- Nucleotide and deduced amino acid sequence relatedness.
- Antigenic relatedness.
- Host of origin.
Pestivirus species demarcation considers several parameters and their relationship to the type viruses of the four currently recognized species (BVDV-1 NADL; BVDV-2 890; BDV X818; and CSFV A187). Nucleotide sequence relatedness is an important criterion for pestivirus species demarcation. For example, the 5′-NCR sequences among the four currently recognized species are over 15% divergent. In most cases, the degree of genetic variation within the 5′-NCR will allow species demarcation. However, in some cases the nt sequence relatedness may be ambiguous and must be complemented with additional comparative analyses. Convalescent animal sera generated against members of a given species (e.g., Bovine viral diarrhea virus 1) generally show a several-fold higher neutralization titer against viruses of the same species than against viruses from the other species. Finally, differences in host of origin and disease can assist in species identification.
For example, Bovine viral diarrhea virus 1 and Classical swine fever virus are considered different species because their members differ from each other by: (i) at least 25% at the sequence level (complete genomes), (ii) at least 10-fold difference in neutralization titer in cross-neutralization tests with polyclonal immune sera, and (iii) host range, in that under natural conditions CSFV infects only pigs while BVDV-1 infects ruminants as well as pigs.
The two species BVDV-1 and BVDV-2 are often referred to as genotypes 1 and 2 of BVDV. The genetic and antigenic differences between BVDV-1 and BVDV-2 are comparable to the ones among isolates of species BDV, which has been classified into several genotypes (e.g. BDV-1, -2, etc). For CSFV three genotypes (CSFV-1, CSFV-2, and CSFV-3) are recognized. In addition, the genotypes of these Pestivirus species can be further divided into subgroups.
In order to officially establish a novel pestivirus species or genotype, the complete genomic sequence of at least one virus isolate together with data on antigenic relatedness should be provided.
List of species in the genus Pestivirus
| Border disease virus |
|
|
| Border disease virus 1a BD31 | [U70263] | (BDV-1 BD31) |
| Border disease virus 1a X818 | [AF037405] | (BDV-1 X818) |
| Border disease virus 2 Reindeer-1 | [AF144618] | (BDV-2 V60) |
| Border disease virus 3 Gifhorn | [GQ902940] | (BDV-3 Gifhorn) |
| Border disease virus 4 Chamois-1 | [GU270877] | (BDV-4 H2121) |
| Bovine viral diarrhea virus 1 |
|
|
| Bovine viral diarrhea virus 1a NADL | [M31182] | (BVDV-1a NADL) |
| Bovine viral diarrhea virus 1a SD1 | [M96751] | (BVDV-1a SD1) |
| Bovine viral diarrhea virus 1b CP7 | [U63479] | (BVDV-1b CP7) |
| Bovine viral diarrhea virus 1b Osloss | [M96687] | (BVDV-1b Osloss) |
| Bovine viral diarrhea virus 2 |
|
|
| Bovine viral diarrhea virus 2 C413 | [AF002227] | (BVDV-2 C413) |
| Bovine viral diarrhea virus 2 NewYork’93 | [AF502399] | (BVDV-2 NY93) |
| Bovine viral diarrhea virus 2 890 | [U18059] | (BVDV-2 890) |
| Classical swine fever virus |
|
|
| (Hog cholera virus) |
|
|
| Classical swine fever virus 1.1 Alfort/187 | [X87939] | (CSFV-1.1 A187) |
| Classical swine fever virus 1.1 C-strain | [Z46258] | (CSFV-1.1 C) |
| Classical swine fever virus 1.2 Brescia | [M31768] | (CSFV-1.2 Brescia) |
| Classical swine fever virus 2.3 Alfort-Tübingen | [J04358] | (CSFV-2.3 Alfort-T) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses that may be members of the genus Pestivirus but have not been approved as species
| Atypical pestivirus | [FJ040215] | Th/04_KhonKaen |
| Bungowannah virus | [EF100713] | Bungo |
| Giraffe-1 pestivirus | [AF144617] | Giraffe-1 (H138) |
| Pronghorn antelope pestivirus | [AY781152] | Pronghorn |
Genus Hepacivirus
Type species Hepatitis C virus
Distinguishing features
Hepatitis C virus (HCV) is transmitted between humans, principally via exposure to contaminated blood or blood products. There is no known invertebrate vector. Hepaciviruses differ from members of the genera Flavivirus and Pestivirus by their limited ability to be propagated in cell culture, since only a single HCV strain, JFH1, has been found to efficiently infect cultured cells, and only one specific human hepatoma cell line (Huh7) is susceptible to infection with this strain. In the HCV precursor protein, the NS2-3 junction is auto-catalytically cleaved by Zn-dependent NS2-3 protease activity.
Virion properties
Morphology
Virions are about 50 nm in diameter, as determined by filtration and electron microscopy. They are spherical in shape and contain a lipid envelope, as determined by electron microscopy and inactivation by chloroform. The viral core is spherical and about 30 nm in diameter. Detailed structural properties have not been determined.
Physicochemical and physical properties
Virion Mr has not been determined. Buoyant density in sucrose is predominantly about 1.06 g cm−3 for virus recovered from serum during acute infections while more dense forms (ca. 1.15–1.18 g cm−3) predominate when recovered from the serum of chronically infected individuals. The lower density results from physical association of the virion with serum very-low-density lipoproteins (VLDLs). The higher density results from the binding of serum antibodies to the virion. A buoyant density range in isosmotic iodixanol gradients of 1.01–1.10 g cm−3 has been measured for HCV recovered from hepatoma cells infected with HCV; the different densities are believed to be due to differences in the association with low-density lipoproteins/VLDL components. The S20,w is equal to or greater than 150S. The virus is stable in buffer at pH 8.0–8.7. Virions are sensitive to heat, organic solvents and detergents.
Nucleic acid
Virions of HCV contain a single positive sense, infectious ssRNA (Figure 5). The genome length is about 9.6 kb. The 5′-NCR contains an IRES and is about 340 nt long. The 3′-NCR contains a sequence-variable region of about 50 nt, a polypyrimidine-rich region averaging about 100 nt (but highly variable in length) and a highly conserved 98 nt long 3′-terminal region with three defined stem-loop RNA secondary structures. There are at least two seed sites in the HCV 5′-NCR for the liver abundant microRNA miR-122 that are required for efficient HCV replication.
A divergent strain of the genus named “GB virus B” has a closely similar genome organization to HCV. However, it has a longer 5′-NCR of 445 nt with an HCV-like IRES structure. Furthermore, the RNA encodes a shorter polyprotein of 2864 aa, and with a substantial sequence divergence to HCV, with only 28% aa identity between the encoded polyproteins, compared to within the Hepatitis C virus species itself (>60% aa identity).
Proteins
The HCV virion consists of at least three proteins: the nucleocapsid core protein C (p19-21), and two envelope glycoproteins, E1 (gp31) and E2 (gp70). An additional protein, p7 (believed to have properties of an ion channel protein important in viral assembly) is incompletely cleaved from a precursor of E2 to yield E2-p7 and p7, but it is not known whether these are virion structural components. In GB virus B, there is a corresponding protein, p13, that apparently can be cleaved into p7 and p6 proteins. The two envelope glycoproteins can associate as non-covalent heterodimers; recent data, however, indicate that they are covalently linked complexes in virions. The recognized nonstructural proteins include NS2 (21 kDa protein that, before cleavage, is part of a Zn-dependent cysteine protease that bridges NS2 and NS3 and mediates autocatalytic cleavage of the NS2/NS3 junction, and is involved with virus assembly and release), NS3 (70 kDa protein with additional serine protease, helicase and NTPase acitivities; the NS3 protease cleaves the remaining junctions between nonstructural proteins), NS4A (6 kDa cofactor essential for trans NS3 serine protease activity), NS4B (27 kDa protein that induces a membranous replication complex at the endoplasmic reticulum), NS5A (a serine phosphoprotein of unknown specific function, but critical for viral replication and assembly, that exists in 56 and 58 kDa forms, depending on the degree of phosphorylation) and NS5B (68 kDa protein with RdRp activity).
Lipids
Virions have a lipid bi-layer envelope. Historically, based on observed removal of the viral envelope and loss of infectivity following exposure to solvents or detergents, the presence of lipids was inferred. Recently, it has become apparent that the host lipid metabolism plays a critical role in the viral life cycle.
Carbohydrates
The E1 and E2 glycoproteins contain numerous N-linked glycosylation sites, and the demonstration of carbohydrate associated with the products of these two HCV genes expressed as recombinant proteins or in HCV retroviral pseudo-particles is consistent with the presence of carbohydrates in virions. E1 and E2 are TM type I glycoproteins, with C terminal ER retention signals, anchored within the lumen of the endoplasmic reticulum. They are apparently masked when budding occurs allowing the virion to move through the secretory pathway. Recent data in culture systems indicate that N-linked glycans of E1 remain in the high-mannose chains lacking complex carbohydrate, whereas those of E2 are modified. Glucosylation influences E1–E2 heterodimer formation, folding and assembly and release of virions.
Genome organization and replication
The genome contains a single large ORF encoding a polyprotein of about 3000 aa (Figure 5). The gene order is 5′-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-3′. All three structural proteins (C, E1, E2) are encoded within the amino-terminal portion of the large ORF. Immediately downstream is a small protein, p7 (HCV) or p13 (GB virus B), followed by the nonstructural proteins in the 3′ portion of the ORF. Replication is poorly understood but occurs in association with intracytoplasmic membranes. Replicative forms of viral RNA have been detected in liver tissue. The genomic RNA is translated into a polyprotein that is rapidly processed both co- and post-translationally by host and viral proteases. Translation initiation occurs via an IRES within the 5′-NCR, which also contains several closely spaced AUGs. Translocation of the structural glycoproteins to the endoplasmic reticulum probably occurs via internal signal sequences. Cleavage of the structural proteins is mediated by host cell signal peptidases, and signal peptide peptidase. With the exception of the p7/NS2 signalase cleavage, viral proteases cleave all non-structural protein junctions. Virus assembly is believed to occur by budding into vesicles from the endoplasmic reticulum.
Antigenic properties
Virus-specific antibodies to recombinant-expressed structural proteins (C, E1 and E2) and nonstructural proteins (principally NS3, NS4 and NS5) have been detected in individuals infected with HCV. Both linear and conformational epitopes are believed to be involved in the humoral immune response of the host to infection. Significant antigenic diversity throughout the genome is reflected in heterogeneity in the humoral immune response, especially to the product of the NS4 gene. In HCV, high variability is found in the N-terminal 27 aa of E2 (hypervariable region 1; HVR1). The HVR1 contains an HCV neutralization epitope and escape variants of HVR1 are positively selected by the host humoral immune response. Other neutralization epitopes have been identified in E2 outside of HVR1, and at least one neutralization epitope has been identified in E1. Cell-mediated immune responses to all HCV proteins have been detected; it is believed that such responses are associated with amelioration or resolution of infection. With the recent development of the JFH1 culture system, and of intra- and intergenotypic genotype 1-7 JFH1-based recombinant viruses with strain-specific structural proteins, it is now possible to carry out in vitro virus neutralization assays.
Biological properties
Host range
Humans are the natural host and apparent reservoir of hepatitis C virus. The virus can be transmitted experimentally to chimpanzees. No other natural host has been identified. The natural host for GB virus B is not known.
Transmission
Hepatitis C virus is transmitted almost exclusively by parenteral exposure to blood, blood products and objects contaminated with blood. Effective screening of blood donors and implementation of inactivation procedures have virtually eliminated the transmission of HCV via blood and blood products, but other routes of exposure, principally via blood-contaminated syringes, are now the most important recognized risk factors in developed countries. Sexual and mother-to-child transmission has been documented, but are relatively uncommon.
Geographical distribution
HCV has a worldwide distribution. Antibody-based epidemiological studies suggest that about 0.1–2% of the populations of developed countries may be infected with HCV, but antibody prevalence as high as 20% has been detected in some developing countries. The high prevalence of antibody to HCV is thought to be the result of using contaminated needles and syringes in such countries. Most estimates suggest that about 3% of the world population has been infected with HCV, and that more than 150 million people are chronically infected. There are about 4 million new infections per year.
Pathogenicity
Infections range from subclinical to acute and chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. Persistence of the virus occurs in 60–80% of HCV infections, depending on the population studied. In about 20% of the cases, persistent HCV will progress to chronic active hepatitis and cirrhosis, usually over the course of many years. Patients with liver cirrhosis have an approximately 5% risk per year of developing hepatocellular carcinoma.
Persistent HCV infection has been epidemiologically linked to primary liver cancer, cryptogenic cirrhosis and some forms of autoimmune hepatitis. Extrahepatic manifestations of HCV infection include mixed cryoglobulinemia with associated membrano-proliferative glomerulonephritis and, possibly, porphyria cutanea tarda, Sjögren’s-like syndromes and other autoimmune conditions.
Like HCV, GB virus B causes hepatitis and replicates in the liver. However, it only infects tamarins and owl monkeys, not humans or chimpanzees. GB virus B causes self-limiting hepatitis in experimentally infected tamarins and owl monkeys, whereas HCV typically causes chronic hepatitis in man and chimpanzees. Only one variant of GB virus B has been identified to date, in contrast to hundreds of HCV variants.
Cell tropism
HCV has been reported to replicate in several cell lines derived from hepatocytes and lymphocytes, but virus growth has only been sufficient for practical application of these systems in a human hepatoma cell line, Huh7 cells and derivatives. In vivo, HCV replicates in hepatocytes and possibly lymphocytes.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Hepacivirus
The genus Hepacivirus comprises a single species, Hepatitis C virus. However, the species can be classified into seven genetic groups (termed genotypes; see table below), based upon the genome-wide heterogeneity of isolates recovered throughout the world. These differ from each other by about 30–35% at the nt level. Within each genotype, there are a number of subtypes, differing from each other by about 15–25% at the nt level. Although genotypes are distinct genetically, discrimination of subtypes is less clear, particularly in areas of high diversity such as sub-Saharan Africa and South-East Asia. Because systematic serological typing by virus neutralization has not been performed to date, and because major genotypes do not have any other taxonomic characteristics except, in some cases, geographic distribution and differences in treatment response, the seven genetic groups of HCV currently comprise one species. Complete or near complete genomic sequences have been obtained from each of the seven genotypes of HCV, and functionality (replication competence) has been demonstrated by intrahepatic chimpanzee inoculation of RNA transcripts from cDNA clones of genotypes 1a [strains H77 (AF011751 and AF009606), HCV-1 (AF271632), HC-TN (EF62148)], 1b [Con1 (AJ238799), HCV-N (AF139594)], 2a [HC-J6 (AF177036) and JFH1 (AB047639)], 3a [S52 (GU814264)] and 4a [ED43 (GU814264)].
| Hepatitis C virus |
|
|
| HCV genotype 1a /HPCPLYPRE | [M62321] | (HCV-1) |
| HCV genotype 1b /HPCJCG | [D90208] | (J) |
| HCV genotype 2a /HPCPOLP | [D00944] | (J6) |
| HCV genotype 2b /HPCJ8G | [D01221] | (J8) |
| HCV genotype 3a /HPCEGS | [D17763] | (NZL-1) |
| HCV genotype 3k /HPCJK049E1 | [D63821] | (JK049) |
| HCV genotype 4a /HCV4APOLY | [Y11604] | (ED43) |
| HCV genotype 4d | [DQ516083] | (24) |
| HCV genotype 5a /HCV1480 | [Y13184] | (HCV1480) |
| HCV genotype 6a /HCV12083 | [Y12083] | (EUHK2) |
| HCV genotype 6g /HPCJK046E2 | [D63822] | (JK046) |
| HCV genotype 7a | [EF108306] | (QC-69) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses that may be members of the genus Hepacivirus but have not been approved as species
GB virus B comprises one isolate, and its functionality (replication competence) has been demonstrated by intrahepatic inoculation of RNA transcripts from a cDNA clone into tamarins (AF179612).
| GB virus B | [U22304] | (GBV-B) |
Phylogenetic relationships within the family
Divergent members of the family Flaviviridae: evidence for a possible fourth genus
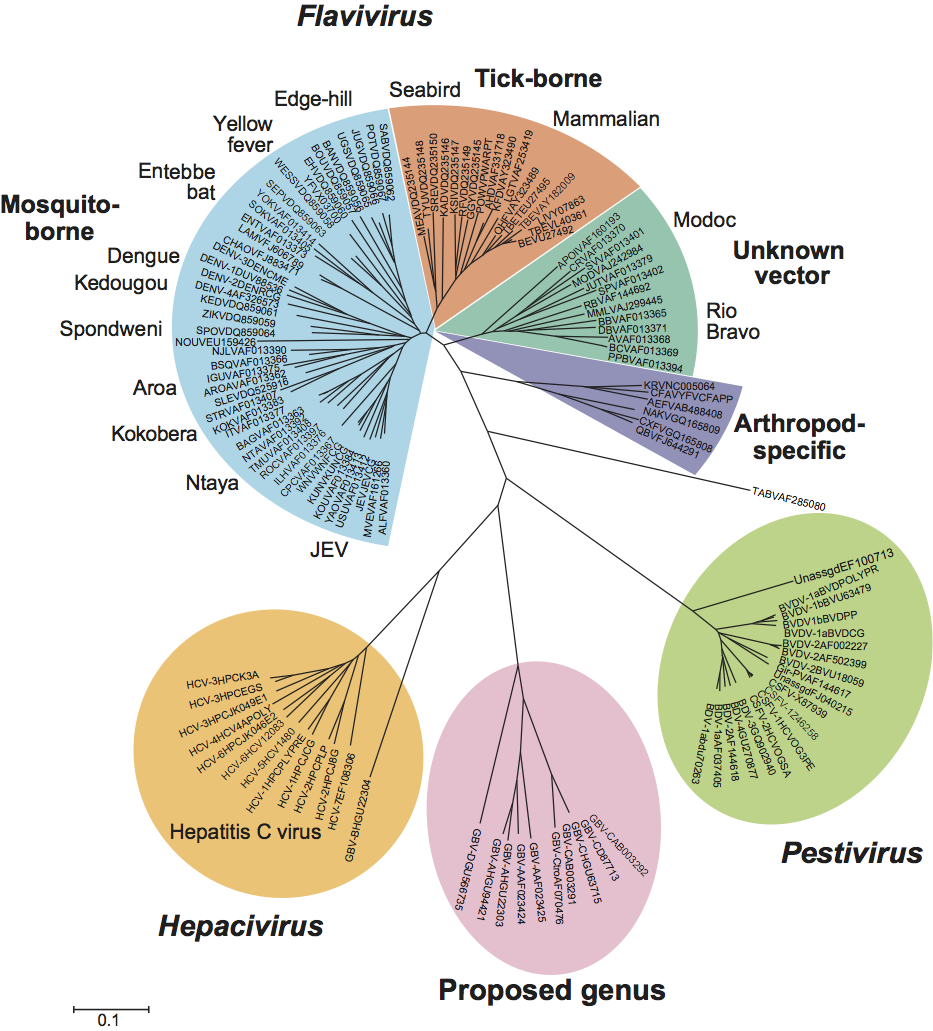
Viruses identified in humans (GBV-C, also known as hepatitis G virus; HGV), chimpanzees (GBV-Ctro), a range of New World monkey species (GBV-A), and a fruit bat (GBV-D), show distant sequence relatedness to other members of the family Flaviviridae, forming a distinct cluster based on phylogenetic analysis of RdRp (Figure 6) and helicase sequences. In addition to their separate phylogenetic position, they show several differences in genome organization from Hepacivirus and other Flaviviridae genera (including an IRES structurally unrelated to those of hepaciviruses and pestiviruses, and the apparent absence of a gene encoding a nucleocapsid protein). These differences merit consideration of this cluster as a separate genus.
GBV-A and GBV-A-like agents are a group of related viruses that have been identified in at least six species of New World monkeys. They do not cause hepatitis in the unique host species of each virus nor in other susceptible species. Their organ site of replication has not been identified and, although the viruses are transmissible via blood, their natural route of transmission is unknown. They cause persistent infection in their respective host species.
GBV-C (or HGV) infects humans worldwide, as well as chimpanzees and potentially other apes. In both humans and chimpanzees, frequencies of viremia range from 2 to 10%. Although GBV-C is transmitted via blood and blood products, its primary route of infection is through sexual contact and from mother-to-child. GBV-C establishes persistent infections in a proportion of those infected (<25%), recovery being associated with seroconversion for anti-E2 antibodies (viremic individuals remain seronegative). Although originally described as a hepatitis virus, it rarely, if ever, causes hepatitis, and its pathogenicity and organ site of replication remain controversial. Some studies suggested that lymphocytes might be its primary site of replication.
GBV-D is the most genetically divergent member of this group. It was detected and genetically characterized from sera from an Old World frugivorous bat (Pteropus giganteus). Infected bats similarly show no evidence of hepatitis, while its frequent detection in serum of this bat species indicates likely persistence of infection.
These viruses show greatest similarity in overall genomic organization and sequence relatedness to hepaciviruses (Figure 6), but differ in that they appear to lack a complete nucleocapsid protein gene, they lack the type 3 IRES found in hepaciviruses (including GBV-B) and pestiviruses, and their 3′-NCR is less complex than that of the hepaciviruses.
The names currently assigned to viruses in this putative genus are regarded as unsuitable for several reasons, being based on a patient’s name (initials GB) or an unproven disease association (HGV). The letter suffixes (-A, -C and -D) do not indicate host origin or genetic relationships. Proposals for a revision of their nomenclature and assignment of a fourth genus in the family Flaviviridae are under consideration.
| GBV-A | [U94421] | (Alab) |
| GBV-C | [U63715] | (GBV-EA) |
| GBV-C | [U45966] | (HGV-R10291) |
| GBV-C(chimpanzee) | [AF070476] | (GBV-Ctro) |
| GBV-D | [GU566735] | (GBV-D strain 93) |
Viruses in the divergent group of GB flaviviruses, members of a possible fourth genus
Similarity with other taxa
The RdRp of flaviviruses shows distant similarity with those of some plant virus families (e.g. Tombusviridae) and has been assigned into RNA virus supergroup 3. However, virion structure and other viral structural and nonstructural genes are distinct and likely non-homologous.
Derivation of names
Flavi: from Latin flavus, ”yellow”.
Pesti: from Latin pestis, “plague”.
Hepaci: from Greek hepar, hepatos, “liver”.
Further reading
Gottwein et al., 2010 J.M. Gottwein, T.K.H. Scheel, B. Callendret, Y-P. Li, H.B. Eccleston, R.E. Engle, S. Govindarajan, W. Satterfield, R.H. Purcell, C.M. Walker, J. Bukh, Novel infectious cDNA clones of hepatitis C virus genotype 3a (Strain S52) and 4a (Strain ED43): genetic analyses and in vivo pathogenesis studies. J. Virol. 84 (2010) 5277–5293.
Grard et al., 2010 G. Grard, G. Moureau, R.N. Charrel, E.C. Holmes, E.A. Gould, X. de Lamballerie, Genomics and evolution of Aedes-borne flaviviruses. J. Gen. Virol. 91 (2010) 87–94.
Gubler et al., 2007 D.J. Gubler, G. Kuno, L. Markoff, D.M. Knipe, P.M. Howley, FlavivirusesFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott Williams and Wilkins, Philadelphia20071153–1252.
Junglen et al., 2009 S. Junglen, A. Kopp, A. Kurth, G. Pauli, H. Ellerbrok, F.H. Leendertz, A new flavivirus and a new vector: characterization of a novel flavivirus isolated from uranotaenia mosquitoes from a tropical rain forest. J Virol. 83 (2009) 4462–4468.
Lemon et al., 2007 S.M. Lemon, C. Walker, Yi, M. Alter, D.M. Knipe, P.M. Howley, Hepatitis C virusFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott Williams and Wilkins, Philadelphia20071253–1304.
Lindenbach et al., 2007 B.D. Lindenbach, H.-J. Thiel, C.M. Rice, D.M. Knipe, P.M. Howley, Flaviviridae: The viruses and their replicationFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott Williams and Wilkins, Philadelphia20071101–1152.
Meyers and Thiel, 1996 G. Meyers, H.-J. Thiel, Molecular characterization of pestiviruses. Adv. Virus Res. 47 (1996) 53–118.
Mukhopadhyay et al., 2005 S. Mukhopadhyay, R.J. Kuhn, M.G. Rossmann, A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3 (2005) 13–22.
Simmonds et al., 2005 P. Simmonds, J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D.G. Murphy, H. Okamoto, J.M. Pawlotsky, F. Penin, E. Sablon, I.T. Shin, L.J. Stuyver, H.-J. Thiel, S. Viazov, A.J. Weiner, A. Widell, Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 42 (2005) 962–973.
Stapleton et al., 2011 J. Stapleton, S. Foung, A.S. Muerhoff, J. Bukh, P. Simmonds, The GB viruses: a review and proposed re-classification of GBV-A, GBV-C (HGV), and GBV-D as pegiviruses. J. Gen. Virol. 92 (2011) 233–246.
Vilcek et al., 2010 S. Vilcek, K. Willoughby, P. Nettleton, P. Becher, Complete genomic sequence of a border disease virus isolated from Pyrenean chamois. Virus Res. 152 (2010) 164–168.
Contributed by
Simmonds, P., Becher, P., Collett, M.S., Gould, E.A., Heinz, F.X., Meyers, G., Monath, T., Pletnev, A., Rice, C.M., Stiasny, K., Thiel, H.-J., Weiner, A. and Bukh, J.
The chapter in the Eighth ICTV Report, which served as the template for this chapter, was contributed by Thiel, H.-J., Collett, M.S., Gould, E.A., Heinz, F.X., Houghton, M., Meyers, G., Purcell, R.H. and Rice, C.M.
Figures
Figure 1 Three-dimensional cryo-electron microscopic reconstructions of immature (left) and mature (right) particles of an isolate of dengue virus (courtesy of M. Rossmann). Shown is a surface rendering of immature dengue virus at 12.5A resolution (left) and mature DENV at 10A resolution (right). The viruses are depicted to scale, but not colored to scale. Triangles outline one icosahedral unit.
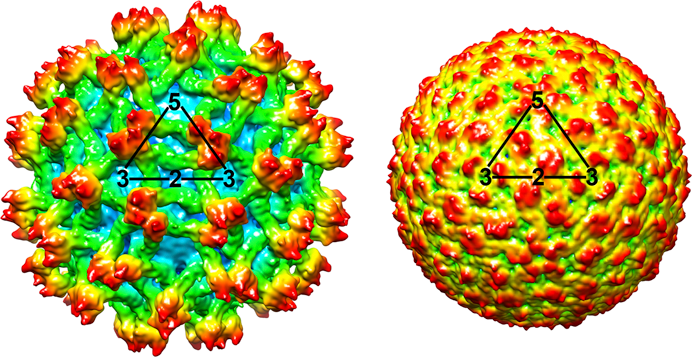
Figure 2 Flavivirus genome organization (not to scale) and polyprotein processing. The virion RNA is about 11 kb in size. At the top is the viral genome with the structural and nonstructural protein coding regions and the 5- and 3-NCRs. Boxes below the genome indicate viral proteins generated by the proteolytic processing cascade. P, H, and R symbols indicate the localization of the NS3 protease, the NS3 RNA helicase, and the NS5 RdRp, respectively.
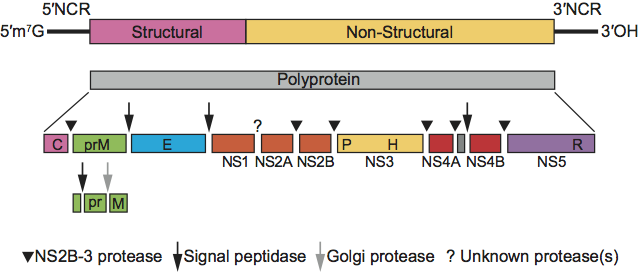
Figure 3 Negative contrast electron micrograph of particles of an isolate of bovine viral diarrhea virus 1. The bar represents 100 nm.
(From M. Knig, with permission.)

Figure 4 Pestivirus genome organization (not to scale) and polyprotein processing. The RNA is usually about 12.3 kb in size (depending on the virus). The 5-NCR is about 370385 nt, the ORF about 11.7 kb and the 3-NCR is 185-273 nt. The viral nonstructural proteins are indicated as NS. P, P, P, H and R symbols indicate the localization of the Npro protease, the NS2 protease, the NS3 protease, the NS3 RNA helicase and the NS5B RdRp, respectively. The proteases and proteolytic steps involved in the generation of individual proteins are indicated. In noncp BVD viruses, NS2-3 cleavage is detectable only for a short time early after infection. In cp BVD viruses, NS3 is produced continuously in addition to NS2-3.
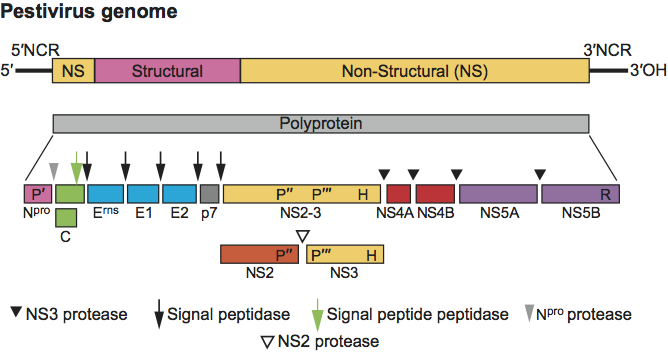
Figure 5 Hepacivirus genome organization (not to scale) and polyprotein processing. For species Hepatitis C virus, the RNA is about 9.6 kb in size. The 5-NCR is about 340 nt, the 3-NCR is about 250 nt, and the ORF is about 9 kb. The species HCV has a p7 protein between E2 and NS2; the other possible member of the genus, GB virus B, has a p13 protein, which can be cleaved into p7 and p6. The host and viral proteases involved in cleavage of the polyprotein are indicated. The cleavage by host signal peptide peptidase (at the C-terminus of core) is indicated by an open arrow; the cleavages by host signal peptidase (remaining sites) are indicated by filled arrows. The locations of the NS2-3 protease, NS3 protease, NS3 RNA helicase and NS5B RdRp are indicated by P, P, H and R, respectively.

Figure 6 Phylogeny of the conserved sequences in the RdRp (NS5 or NS5B) of classified members of the family Flaviviridae. Partial gene sequences (292 aa, positions 77048550 numbered as in the HCV-1 genome, AF011751) from the representative strains listed in each table. CLUSTALW was used to create a multiple alignment for the aa sequences which was verified by alignment of the known motifs in the region. An unrooted phylogenetic tree was constructed from the sequence alignment by neighbor-joining of (uncorrected) amino acid p-distances using the MEGA version 4.1 package. The virus names corresponding to the abbreviations can be found in the List of species in each genus. Note that the Entebbe virus group comprising Entebbe bat virus, Sokoluk virus and Yokose virus have no known vector but group phylogenetically within the mosquito-borne flaviviruses. Nounan, Chaoyang, Ngoye and Lammi viruses are not currently assigned to a specific virus group but also group within the mosquito-borne viruses.