Family: Closteroviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are helically constructed filaments with a pitch of the primary helix in the range of 3.4–3.8 nm, containing about 10 protein subunits per turn of the helix and showing a central hole of 3–4 nm (Figure 1 top). The very flexuous and open structure of the particles is the most conspicuous trait of members of the family. Virions have a diameter of about 12 nm and lengths ranging from 650 nm (species with fragmented genome) to over 2000 nm (species with monopartite genome). The fragility of virions and a tendency to end-to-end aggregation contribute to the fact that a range of lengths is often given for single viruses. Two types of coat proteins, the major CP and a CP analog, or minor CP (CPm), are the most abundant protein components involved in the formation of (most) closterovirid particles. CPm encapsidates the 600–700 5′-terminal nucleotides of the viral RNA, coating one extremity (75–100 nm) of the closterovirid particle, as shown for isolates of beet yellows virus (BYV), carrot yellow leaf virus (CYLV), citrus tristeza virus (CTV), grapevine leafroll-associated virus 2 (GLRaV-2) and lettuce infectious yellows virus (LIYV), thus forming a distinct structure for which the terms “rattlesnake”, “heterodimeric” or “bipolar” have been coined (Figure 1 bottom).
Physicochemical and physical properties
Virions usually sediment as a single band in sucrose or Cs2SO4 gradients. S20,w is around 130–140, buoyant density is 1.33 g cm−3 in CsCl and 1.257 g cm−3 in Cs2SO4. Virions of several species are degraded by CsCl and are unstable in high salt concentration, resist moderately high temperatures (thermal inactivation is around 45–55 °C) and organic solvents, but are sensitive to RNase and chelation.
Nucleic acid
Regardless of the genome type, monopartite or fragmented, virions contain a single molecule of linear, positive sense, single stranded RNA, constituting 5–6% of the particle weight. Genome size is related to particle length, ranging from 13,000 to slightly over 19,000 nucleotides. The 5′ end of the genome is likely to be capped. The 3′ end is not polyadenylated and does not possess a tRNA-like structure, but has several hairpin structures and a putative pseudoknot essential for replication.
Proteins
Structural proteins of most members of the family consist of a major CP and of a diverged copy of it denoted minor CP (CPm), with a size ranging from 22 to 46 kDa (CP) and 23 to 80 kDa (CPm), according to the individual species. A group of ampeloviruses with a small-sized genome (ca. 13,000 nt) apparently lacks a true CPm. With BYV, and presumably for most other members of the family, CPm is required for the assembly of the 5′-extremity of the virion, the protein of about 60 kDa is required for incorporation of both HSP70h and CPm to virions, which also incorporate a 20 kDa protein that may form the tip segment of the virion head.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Members of the family have one of the largest genomes among plant viruses because of sequence duplication and acquisition of nonviral coding sequences (e.g., protease, HSP70 protein) via RNA recombination. Recombination may also explain differences in genome organization between genera and members of the same genus. Genome organization, i.e. the number and relative position of the ORFs differs between the genera and/or individual viral species. However, the complex ORF1a–ORF1b invariably encodes the replication-related proteins, with methyl-transferase (Mtr), helicase (Hel), and RNA-dependent RNA polymerase (RdRp) conserved domains. Downstream ORFs, which encode in 5′→3′ direction a 6K small hydrophobic protein, the HSP70h, the ~60 kDa protein, the CP and CPm, form a five-gene module which is conserved, with few modifications, among most members of the family analysed so far. The HSP70h and the ~60 kDa proteins are integral virion components present in all the sequenced members of the family. The functions postulated for HSP70h are: mediation of cell-to-cell movement through plasmodesmata, involvement in the assembly of multisubunit complexes for genome replication and/or subgenomic RNAs synthesis, and assembly of virus particles. The ~60 kDa protein is required for incorporation of both HSP70h and CPm to virion heads. The duplication of the capsid protein gene seems to be the only example of such condition among viruses with elongated particles. In general, capsid proteins and their homologs (CPm) show a significant degree of sequence conservation and the duplicate copies probably retain the general spatial folding and some crucial properties of the CPs. Notable exception are a group of ampeloviruses with the smallest genomes in the family [e.g. grapevine leafroll-associated virus 4 (GLRaV-4), GLRaV-5, GLRaV-6, GLRaV-9, pineapple mealybug wilt-associated virus 1(PMWaV-1) and PMWaV-3] which do not appear to possess CPm. The genome expression strategy is based on: (i) proteolytic processing of the polyprotein encoded by ORF1a; (ii) +1 ribosomal frameshift for the expression of the RdRp domain encoded by ORF1b, a mechanism not found in other (+)RNA plant viruses; (iii) expression of the downstream ORFs via the formation of a nested set of 3′ co-terminal sub-genomic RNAs (sgRNAs). The dsRNA patterns are very complex and variable among species, reflecting the different numbers and sizes of the ORFs present in individual genomes and, in some cases, the existence of defective RNAs. Replication occurs in the cytoplasm, possibly in association with endoplasmic reticulum-derived membranous vesicles and vesiculated mitochondria. From an evolutionary point of view, closteroviruses represent a monophyletic virus lineage that might have evolved from a smaller filamentous virus when higher plants have differentiated. This progenitor virus, thought to be composed of three genes encoding replication-associated proteins, a protein (p6) with affinity for cell membranes, and a single coat protein, acquired the HSP70h and a ~60 kDa protein derived from a fusion of two domains, N-terminal domain of unknown evolutionary provenance, and a duplicated capsid protein domain. Under the pressure of further modular evolutionary events, i.e. duplication of the coat protein gene, acquisition of diverse suppressors of RNA silencing and of additional genes acquired via horizontal gene transfer (e.g. papain-like cysteine proteinase and AlkB domains), this family ancestor gave rise to the progenitors of the three extant genera of the family. One of these genera (Crinivirus), differentiated further by splitting its genome in two or three genome components.
Antigenic properties
Virion proteins are moderately antigenic. Most virus species within genera are serologically unrelated or distantly related to one another. No intergeneric serological relationship has been detected.
Biological properties
The natural and experimental host ranges of individual virus species are usually restricted, except for a few members of the genus Crinivirus. Disease symptoms are of the yellowing type (i.e. stunting, rolling, yellowing or reddening of the leaves, small and late ripening fruits), or pitting and/or grooving of the woody cylinder of woody hosts. Infection is systemic, but usually limited to the phloem, which may necrotize to a varying extent. Few species of the genus Closterovirus are transmissible by mechanical inoculation, though with difficulty, but none of those in the genera Ampelovirus and Crinivirus. In vegetatively propagated crops long distance virus dissemination is primarily through infected propagating material. Some species can be transmitted through dodder (Cuscuta spp.). Transmission through seeds has not been ultimately proven. According to the genus, natural vectors are aphids, whiteflies, pseudococcid mealybugs and soft scale insects. Transmission is semi-persistent regardless of the type of vector. Geographical distribution varies from restricted to widespread, depending on the virus species, most of which occur in temperate or subtropical regions. Virions are usually found in the phloem (sieve tubes, companion cells, phloem parenchyma), occasionally in the mesophyll and epidermis. Ultrastructural modifications arise by membrane proliferation, degeneration and vesiculation of mitochondria, and formation of inclusion bodies. These are made up of aggregates of virions or membranous vesicles, or a combination of the two. Virions accumulate in conspicuous cross-banded fibrous masses or, more typically, in more or less loose bundles intermingled with single or clustered membranous vesicles. Inclusions of this type are one of the hallmarks of the family. The vesicles contain a fibrillar network and derive either from the endoplasmic reticulum, or from peripheral vesiculation of mitochondria.
Species and genus demarcation criteria within the family
Traits that largely characterize the family and that are the basis of the current classification are:
- Morphology, substructure and size of virus particles.
- Very large, positive sense, single stranded RNA genome with an organization (number and order of genes) quite distinct from those of other plant viruses.
- Possession of unique genes coding for a homolog of the cellular HSP70 heat shock protein (HSP70h) and for a duplicated, diverged copy of the capsid protein (minor protein, CPm).
- Close phylogenetic relationships in replication-related proteins (Mtr, Hel and RdRp).
- Genome expression strategy based on ribosomal frameshift, proteolytic processing and production of subgenomic messenger RNAs.
- Induction of specific cytopathic structures in infected cells, consisting of cytoplasmic aggregates of virus particles intermingled with single or clustered membranous vesicles.
- Specific tissue tropism (members are mostly phloem-limited).
- Natural transmission by aphids, mealybugs or whiteflies in a semi-persistent manner; experimental transmission by mechanical inoculation very difficult or not possible.
Table 1 Distinguishing properties of the genera in the family Closteroviridae
| Genus | Virion length (nm) | RNA species (No.) | Genome size (kb) | ORF (No.) | Replicase (kDa) | HSP70h (kDa) | CP (kDa) | CPm (kDa) | Vector |
| Closterovirus | 1350–2000 | 1 | 14.5–19.3 | 8–12 | 349–367 | 65–67 | 22–25 | 24–27 | Aphids |
| Ampelovirus | 1400–2000 | 1 | 13.0–18.5 | 7–12 | 245–293 | 57–59 | 28–36 | 50–56 | Mealybugs |
| Crinivirus | 750–900 | 2 or 3 | 15.6–17.9 | 9–13 | 267–280 | 62–65 | 28–29 | 53–80 | Whiteflies |
Genus Closterovirus
Type species Beet yellows virus
Distinguishing features
The genus comprises species with particle length above 1200 nm, and monopartite RNA genome, 14.5–19.3 kb in size, in which CPm is located upstream of the CP gene. Natural transmission by aphids.
Virion properties
Morphology
Particle morphology largely conforms to that of other members of the family. Virions are of one size, ranging from 1350 to 2000 nm in length. CTV has also smaller than full-length particles that may encapsidate subgenomic or multiple species of defective RNAs (D-RNA) containing all of the cis-acting sequences required for replication. sgRNAs may be involved in the construction of recombinant D-RNAs.
Physicochemical and physical properties
According to the species, infectivity is inactivated at temperatures between 40 and 55 °C, is retained for 1 to 4 days at room temperature, up to 1 year in frozen sap, 2 years in dried leaf material, 5 years in lyophilized preparations stored at −20 °C, and is destroyed at pH lower than 6. A260/A280 ratio is around 1.20 but some members [BYV, carnation necrotic fleck virus (CNFV), burdock yellows virus (BuYV)] lack tryptophan, which results in a higher ratio (1.4–1.8) for the virions. S20,w ranges from 130 (BYV) to 140 (CTV), buoyant density is 1.33 g cm−3 in CsCl (BYV and CTV) and 1.257 g cm−3 in Cs2SO4 (CTV).
Nucleic acid
Virions contain a single molecule of linear, positive sense, single stranded RNA from 14.5 to 19.3 kb in size. Multiple double stranded RNA (dsRNA) species occur in infected tissues, the largest of which is usually the replicative form of the entire genome. sgRNAs generate a range of smaller dsRNAs. With CTV, the presence of D-RNA makes the dsRNA pattern of virus isolates more complex than that of other members of the genus.
Genome organization and replication
Sequenced members of the genus Closterovirus show three types of genome organization exemplified by BYV (Figure 2), CTV (Figure 3) and BYSV: (i) BYV contains eight ORFs flanked by 5′ and 3′ UTRs of 107 and 181 nt, respectively; (ii) CTV has 12 ORFs and UTRs of 107 nt at the 5′ end and 275 nt at the 3′ end. It differs from the BYV genome in having two papain-like protease domains in ORF1a, an extra 5′ proximal ORF (ORF2) encoding a 33 kDa product with no similarity to any other protein in databases, and two extra 3′ proximal ORFs (ORF9 and ORF11); (iii) BYSV has 10 ORFs and a 3′ UTR 241 nt in size, a length intermediate between that of the BYSV and CTV UTRs. A further difference with the BYV genome rests in the presence of an extra ORF (ORF2) encoding a 30 kDa polypeptide with no similarity to any other protein in databases. This ORF is located downstream of ORF1b, i.e. in the same position as the unrelated CTV ORF2. Thus, the organization of BYSV genomes is intermediate between that of BYV and CTV, suggesting that these three viruses might represent three distinct stages in closterovirus evolution. Non-structural proteins common to all members of the genus are: (i) a large polypeptide (over 300 kDa) containing the conserved domains of papain-like protease (P-Pro), methyltransferase (Mtr), and helicase (Hel); (ii) a ~50 kDa protein with all sequence motifs of viral RdRp of the “alpha-like” supergroup of positive-strand RNA viruses; (iii) a 6 kDa hydrophobic protein with membrane-binding properties; (iv) the homolog of the cellular HSP70 heat-shock proteins (HSP70h); (v) a 55–64 kDa product, referred to as the ~60 kDa protein. Some of the structural and non structural proteins function as suppressors of the RNA silencing plant defence machinery. For instance, CP, p20 and p23 proteins of CTV have suppressor activity, much the same as the homologs of p21 of BYSV, BYV, and GLRaV-2. CTV p23 is a unique protein in the family and has a nucleolar localization. Silencing suppressors contribute to the accumulation of virus particles and are important determinants of pathogenesis.
Antigenic properties
No serological relationships reported among different virus species of the genus. Monoclonal antibodies have been produced to BYV, CTV and GLRaV-2 and polyclonal antisera have been raised to BYV, CTV and CYLV from fusion proteins obtained in bacterial expression systems. Polyclonal antisera have been raised to normal capsid proteins of BYV, BYSV, GLRaV-2 and BuYV.
Biological properties
Most of the members of the genus infect herbaceous hosts (weeds, vegetable and flower crops) or shrubs (raspberry). Notable exceptions are CTV, GLRaV-2 and a couple of fig viruses [fig leaf mottle-associated virus 1 (FLMaV1) and fig mild mottle virus (FMMV)] which infect woody hosts. Symptoms are primarily of the yellowing type (rolling, yellowing or reddening of the leaves). Some CTV strains induce stem pitting. Transmission is by aphids with a semipersistent modality. The range of vectors varies for individual viruses from rather wide to restricted. For instance, BYV is transmitted by 23 aphid species (Myzus persicae and Aphis fabae being the main natural vectors), CTV by seven species (Toxoptera citricida and Aphis gossypii being the most efficient vectors) and a number of other viruses [e.g. CNFV, WYLV, BuYV, mint virus 1 (MV-1)] by a single aphid species, or do not have known vectors (e.g. GLRaV-2). Some species can be transmitted by inoculation of sap, though with difficulty (e.g. CTV, GLRaV-2, BYV), but none is transmitted through seeds. Species infecting vegetatively propagated hosts (citrus, grapevine, raspberry) are transmitted by grafting and disseminated over long distances with propagating material. Geographic distribution ranges from very wide (e.g. CTV, GLRaV-2, BYV) to restricted (e.g. BuYV, MV-1, WYLV). The membranous vesicles with a fibrillar content derive from the endoplasmic reticulum.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Particle size.
- Size of CP, as determined by deduced amino acid sequence data.
- Serological specificity using discriminatory monoclonal or polyclonal antibodies.
- Genome structure and organization (number and relative location of the ORFs).
- Amino acid sequence of relevant gene products (polymerase, CP, HSP70h) differing by more than 25%.
- Vector species and specificity.
- Magnitude and specificity of natural and experimental host range.
- Cytopathological features (i.e., aspect of inclusion bodies and origin of cytoplasmic vesicles).
List of species in the genus Closterovirus
| Beet yellow stunt virus |
|
|
| Beet yellow stunt virus-California | [U51931*] | (BYSV-CA) |
| Beet yellows virus |
|
|
| Beet yellows virus-4 | [AF190581] | (BYV-4) |
| Burdock yellows virus |
|
|
| Burdock yellows virus-Japan |
| (BuYV-JA) |
| Carnation necrotic fleck virus |
|
|
| Carnation necrotic fleck virus-Yunnan | [EU884443*] | (CNFV-YN) |
| Carrot yellow leaf virus |
|
|
| Carrot yellow leaf virus-Germany | [FJ869862=NC_013007] | (CYLV-DE) |
| (Heracleum virus 6) |
|
|
| (Hogweed virus 6) |
|
|
| Citrus tristeza virus |
|
|
| Citrus tristeza virus strain T30 | [AF260651] | (CTV-T30, mild isolate) |
| Citrus tristeza virus strain, T36 | [U16304=NC_001661] | (CTV-T36, intermediate severity) |
| Citrus tristeza virus strain VT | [U56902] | (CTV-VT, severe isolate) |
| Grapevine leafroll-associated virus 2 |
|
|
| (Grapevine rootstock stem lesion-associated virus) | [AF314061=NC_004724] | (GRSLaV) |
| Grapevine leafroll-associated virus 2 - PN | [AY881628=NC_007448] | (GLRaV-2-PN) |
| Mint virus 1 |
|
|
| Mint virus 1-454.004 | [AY792620=NC_006944] | (MV-1-454.004) |
| Wheat yellow leaf virus |
|
|
| Wheat yellow leaf virus-Japan |
| (WYLV-JA) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Closterovirus but have not been approved as species
| Alligatorweed stunting virus |
| (AWSV) |
| Clover yellows virus |
| (CYV) |
| Dendrobium vein necrosis virus |
| (DVNV) |
| Festuca necrosis virus |
| (FNV) |
| Fig leaf mottle-associated virus 1 | [AM279676*] | (FLMaV-1) |
| Fig mild mottle virus | [FJ611959*] | (FMMV) |
| Raspberry leaf mottle virus | [DQ357218=NC_008585] | (RLMV) |
| Strawberry chlorotic fleck-associated virus | [DQ860839=NC_008366] | (SCFaV) |
* Sequences do not comprise the complete genome.
Genus Ampelovirus
Type species Grapevine leafroll-associated virus 3
Distinguishing features
The genus comprises species with particles 1400–2000 nm long, monopartite genome 13.0–18.5 kb in size, transmitted by pseudococcid mealybugs and soft scale insects.
Virion properties
Morphology
Particle morphology largely conforms to that of other members of the family.
Physicochemical and physical properties
Information is very limited, except for the size of CP and CPm, as deduced from sequence data.
Nucleic acid
Virions contain a single molecule of linear, positive sense, single stranded RNA from 13.0 to 18.5 kb in size. Multiple double stranded RNA (dsRNA) species occur in infected tissues, the largest of which is usually the replicative form of the entire genome. Smaller dsRNA are thought to be replicative forms of subgenomic RNAs.
Genome organization and replication
The genus Ampelovirus shows a wide variation in genome size and organization. At one extreme there are grapevine leafroll-associated virus 1 (GLRaV-1) and GLRaV-3, which has the largest genome of all (18,498 nt). GLRaV-3 has 12 ORFs, coding for the replication related proteins (ORFs 1a and 1b), two small hydrophobic proteins (6 kDa), the HSP70h, the ~60–kDa protein, CP, CPm and five additional proteins 21, 20, 20, 4 and 7 kDa in size, respectively (Figure 4). The 5’ UTR and 3’ UTR are 737 and 277 nt in size, respectively. GLRaV-1 differs from all other members of the genus in encoding two copies of the CPm. At the other extreme there is a group of viruses infecting grapevine [e.g, grapevine leafroll-associated viruses 5 and 9 (GLRaV-5 and -9)] and pineapple [e.g. pineapple mealybug wilt-associated viruses 1 and 3 (PMWaV-1 and -3)]. All these viruses have a genome made up of seven ORFs and lack the CPm. PMWaV-1, a representative of this group, has a genome 13,071 nt in size, beginning with a 535 nt UTR at the 5′ end, followed by the ORFs expressing, respectively, the replication related proteins, a 6 kDa hydrophobic protein, the HSP70h, the ~60 kDa protein, the CP and a 24 kDa protein. A UTR 132 nt in size terminates the genome. Replication occurs in the cytoplasm, likely in association with membranous vesicles, derived either from the endoplasmic reticulum or from peripheral vesiculation and disruption of mitochondria (GLRaV-1, GLRaV-3). Structural and non-structural proteins are similar in type and function to those reported for the genus Closterovirus.
Antigenic properties
Polyclonal antisera and monoclonal antibodies have been raised to most of the members of the genus. A recombinant single-chain variable fragment antibody was synthesized to GLRaV-3. GLRaV-1 and GLRaV-3 are distantly serologically related based on cross-reactivity to a monoclonal antibody to GLRaV-1. GLRaV-4, -5, -6 and -9 show serological interrelations when tested with polyclonal antisera or monoclonal antibodies. Grapevine leafroll-associated virus 7 (GLRaV-7), an unassigned member of the family, is also distantly related to the four above species. The three pineapple mealybug wilt-associated viruses are serologically unrelated to one another.
Biological properties
The majority of extant ampelovirus species are recorded from woody hosts (grapevine, plum, fig) and pineapple. According to the host, they induce rolling yellowing and reddening of the leaves (grapevine), stem pitting (plum), wilting or symptomless infections (pineapple). Natural vectors are mealybugs which transmit with a semipersistent modality. The range of vectors varies for individual viruses from rather wide to restricted. For instance, GLRaV-1 is transmitted by species of several genera of pseudococcid mealybugs (Heliococcus, Phenacoccus, Pseudococcus) and soft scale insects (Pulvinaria, Neopulvinaria and Parthenolecanium); GLRaV-3 by pseudococcid mealybugs (Planococcus, Pseudococcus, Heliococcus, Phenacoccus) and soft scale insects (Pulvinaria, Neopulvinaria, Parthenolecnium, Coccus, Saissetia, Parasaissetia and Ceroplastes), whereas GLRaV-5 is transmitted by Pseudoccocus, Planococcus and Ceroplastes spp. Vectors of pineapple ampeloviruses are two species of the genus Dysmicoccus, and LChV-2 is transmitted by Phenacoccus aceris. None of the viruses is transmitted through seed or mechanically. All persist in plant parts used for propagation and are disseminated with them over long distances. Geographical distribution is very wide.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Particle size.
- Size of CP, as determined by deduced amino acid sequence data.
- Serological specificity using discriminatory monoclonal or polyclonal antibodies.
- Genome structure and organization (number and relative location and size of the ORFs).
- Amino acid sequence of relevant gene products (polymerase, CP, HSP70h) differing by more than 25%.
- Vector species and specificity.
- Magnitude and specificity of natural and experimental host range.
- Cytopathological features (i.e., aspect of inclusion bodies and origin of cytoplasmic vesicles).
List of species in the genus Ampelovirus
| Grapevine leafroll-associated virus 1 |
|
|
| Grapevine leafroll-associated virus 1-Australia | [AF195822*] | (GLRaV-1-AUS) |
| Grapevine leafroll-associated virus 3 |
|
|
| Grapevine leafroll-associated virus 3-NY1 | [AF037268=NC_004667] | (GLRaV-3-NY1) |
| Grapevine leafroll-associated virus 5 |
|
|
| Grapevine leafroll-associated virus 5-US | [AF233934*] | (GLRaV-5-US) |
| Little cherry virus 2 |
|
|
| Little cherry virus 2-USA6b | [AF531505=NC_005065] | (LChV-2-USA6b) |
| Pineapple mealybug wilt-associated virus 1 |
|
|
| Pineapple mealybug wilt-associated virus 1-Hawaii | [AF414119=NC_010178] | (PMWaV-1-HI) |
| Pineapple mealybug wilt-associated virus 2 |
|
|
| Pineapple mealybug wilt-associated virus 2-Hawaii | [AF283103*] | (PMWaV-2-HI) |
| Pineapple mealybug wilt-associated virus 3 |
|
|
| Pineapple mealybug wilt-associated virus 3-Hawaii | [DQ399259*] | (PMWaV-3-HI) |
| Plum bark necrosis stem pitting-associated virus |
|
|
| Plum bark necrosis stem pitting-associated virus-US | [EF546442=NC_009992] | (PBNSPaV-US) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Ampelovirus but have not been approved as species
| Fig leaf mottle-associated virus 2 | [FJ473383*] | (FLMaV-2) |
| Grapevine leafroll-associated virus 4 | [DQ325516*] | (GLRaV-4) |
| Grapevine leafroll-associated virus 6 | [AJ496796*] | (GLRaV-6) |
| Grapevine leafroll-associated virus 9 | [AY297819*] | (GLRaV-9) |
| Grapevine leafroll-associated Carnelian virus | [FJ907331] | (GLRaV-Car) |
| Grapevine leafroll-associated virus Pr | [AM182328=NC_011702] | (GLRaV-Pr) |
| Sugarcane mild mosaic virus |
| (SMMV) |
* Sequences do not comprise the complete genome.
Genus Crinivirus
Type species Lettuce infectious yellows virus
Distinguishing features
The genus comprises species transmitted by whiteflies. Virions usually have two modal lengths (650–850 and 700–900 nm) and a bipartite genome, but potato yellow vein virus (PYVV) has a tripartite genome.
Virion properties
Morphology
Particle morphology largely conforms to that of other members of the family.
Physicochemical and physical properties
Information is very limited, except for the size of CP and CPm, as deduced from sequence data.
Nucleic acid
Virions contain a single molecule of linear, positive sense, single stranded RNA with size ranging from 7801 to 9127 nt (RNA-1) and from 7903 to 8530 nt (RNA-2) in species with bipartite genome. The RNA size of PYVV, the only species with a tripartite genome, is 8035 nt (RNA-1), 5339 nt (RNA-2) and 3892 nt (RNA-3).
Genome organization and replication
The genome of most criniviruses (e.g. LIYV) is divided between two linear, positive sense, single stranded RNAs totalling 15.6–17.9 kb in size (Figure 5), but PYVV possesses a tripartite genome. All molecules are needed for infectivity and are separately encapsidated. RNA-1 of LIYV contains three ORFs, i.e. the ORF1a–ORF1b complex plus a 3′-most ORF coding for a 32 kDa protein with no similarity to any protein in databases. This ORF is similar in size and location to ORF2 of CTV and BYSV but the respective expression products are not related. RNA-1 has 5′ and 3′ UTRs of 97 and 219 nucleotides, respectively. As with other members of the family, the ORF1a–ORF1b complex codes for the replication-related proteins including the RNA-dependent RNA polymerase (RdRp). RNA-2 has seven ORFs flanked by a 5′ UTR of 326 nt and a 3′ UTR of 187 nt. RNA2 contains the five-gene module which, however, differs from that of members of the genus Closterovirus by the insertion of an extra gene (ORF4) upstream of the CP gene. As to PYVV: (i) RNA-1 (8035 nt in size) is composed of three ORFs, i.e. the ORF1a-ORF1b complex and a 7 kDa hydrophobic protein containing a potential transmembrane helix; (ii) RNA-2 (5,339 nt in size) comprising five predicted ORFs that encode, in the order, the HSP70h; a 7 kDa protein similar to a comparable protein of cucurbit yellow stunting disorder virus (CYSDV); the ~60 kDa protein; a 9.8 kDa product with no significant similarity to any other sequence in database; the 28.2 kDa putative CP; (iii) RNA-3 (3892 nt) has three potential ORFs coding for a protein 4 kDa in size with no counterpart with other proteins in the family and no significant sequence homology in database; the 77.5 kDa CPm, and a 26.4 kDa protein present in other members of the genus. In all criniviruses, the order of the CP and CPm ORFs is reversed compared to that of species in the genus Closterovirus. Sweet potato chlorotic stunt virus (SPCSV) and tomato chlorosis virus (ToCV) have a particularly large CPm (75–80 kDa), compared to LIYV (53 kDa). Replication occurs in the cytoplasm, likely in association with membranous vesicles, derived from the endoplasmic reticulum or from vesiculated mitochondria (CYSDV). Structural and non-structural proteins are similar in type and function to those reported for the genus Closterovirus. Both genomic RNAs of ToCV encode RNA silencing suppressors, e.g. the p22 protein in RNA-1 and CP and CPm in RNA-2. The p25 protein of CYSDV, the viral RNAse III and the p22 gene present in a few isolates of SPCSV also have suppressor activity.
Antigenic properties
Monoclonal antibodies have been produced to proteins of SPCSV. Antisera have been raised from structural and non structural proteins produced as fusion proteins in bacterial expression systems (SPCSV and LIYV) or from CPs [tomato infectious chlorosis virus (TICV), LIYV, lettuce chlorosis virus (LCV) and ToCV]. Generally, there are no detectable interspecific serological relationships. TICV and ToCV, however, are distantly serologically related.
Biological properties
Criniviruses infect primarily herbaceous hosts, in which they induce extensive chlorosis to yellow discoloration of the leaves, often accompanied by stunting. They are transmitted semi-persistently by whiteflies of the genera Trialeurodes and Bemisia. Persistence and specificity of transmission by their respective vectors have been used as characters for species differentiation. Thus, the viruses of group 1 [PYVV, blackberry yellow vein-associated virus (BYVaV), beet pseudoyellows virus (BPYV) and strawberry pallidosis-associated virus (SpaV)] are transmitted by T. vaporariorum, viruses of group 2 [ToCV, SPCSV, CYSDV and bean yellow disorder virus (BYDV)] by B. tabaci, whereas one of the viruses of group 3 is transmitted by B. tabaci (LIYV) and the other by T. vaporariorum (TICV). These groups were identified by comparative phylogenetic analyses of RdRp amino acid sequences. None of the viruses is transmitted through seed or mechanically. Geographical distribution varies from restricted (e.g. BYVaV) to very wide. Some emerging viruses (e.g. CYSDV, TICV and ToCV) are being increasingly recorded from a number of European, American and Asiatic countries. The membranous vesicles with a fibrillar content derive from the endoplasmic reticulum and/or from vesiculated mitochondria (CYSDV).
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Particle size.
- Size of CP, as determined by deduced amino acid sequence data.
- Serological specificity using discriminatory monoclonal or polyclonal antibodies.
- Genome structure and organization (number and relative location of the ORFs).
- Amino acid sequence of relevant gene products (polymerase, CP, HSP70h) differing by more than 25%.
- Vector species and specificity.
- Magnitude and specificity of natural and experimental host range.
- Cytopathological features (i.e., aspect of inclusion bodies and origin of cytoplasmic vesicles).
List of species in the genus Crinivirus
| Abutilon yellows virus |
|
|
| Abutilon yellows virus-California | [AY42270*] | (AbYV-CA) |
| Bean yellow disorder virus |
|
|
| Bean yellow disorder virus-Bn03 | [EU191904=NC_010560, EU191905=NC_010561] | (BYDV-Bn03) |
| Beet pseudoyellows virus |
|
|
| Beet pseudoyellows virus-Maryland | [AY330918=NC_005209, AY330919=NC_005210] | (BPYV-MD) |
| (Cucumber chlorotic spot virus) |
| (CCSV) |
| (Cucumber yellows virus) | [AB085612, AB085613] | (CYV) |
| Blackberry yellow vein-associated virus |
|
|
| Blackberry yellow vein-associated virus-South Carolina | [AY776334=NC_006962, AY776335=NC_006963] | (BYVaV-SC) |
| Cucurbit yellow stunting disorder virus |
|
|
| Cucurbit yellow stunting disorder virus-Spain | [AJ537493, AJ439690] | (CYSDV-ES) |
| Lettuce chlorosis virus |
|
|
| Lettuce chlorosis virus-California | [FJ380118=NC_012909, FJ380119=NC_012910] | (LCV-CA) |
| Lettuce infectious yellows virus |
|
|
| Lettuce infectious yellows virus-California | [U15440=NC_003617, U15441=NC_003618 ] | (LIYV-CA) |
| Potato yellow vein virus |
|
|
| Potato yellow vein virus-Peru | [AJ557128=NC-006061, AJ557129=NC-006062, AJ508757=NC-006063] | (PYVV-Peru) |
| Strawberry pallidosis-associated virus |
|
|
| Strawberry pallidosis-associated virus-M1 | [AY488137=NC_005895, AY488138=NC_005896] | (SpaV-M1) |
| Sweet potato chlorotic stunt virus |
|
|
| Sweet potato chlorotic stunt virus-EA | [AJ428554=NC_004123, AJ428555=NC_004124] | (SPCSV-EA) |
| Tomato chlorosis virus |
|
|
| Tomato chlorosis virus-Florida | [AY903447=NC_007340, AY903448=NC_007341] | (ToCV-FL) |
| Tomato infectious chlorosis virus |
|
|
| Tomato infectious chlorosis virus-California | [FJ815440=NC_013258, FJ814441=NC_013259] | (TICV-CA) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Crinivirus but have not been approved as species
| Diodia vein chlorosis virus | [GQ376201*, GQ225585*] | (DVCV) |
| Cucurbit chlorotic yellows virus | [AB523788, AB523789] | (CCYV) |
* Sequences do not comprise the complete genome.
List of unassigned species in the family Closteroviridae
| Mint vein banding-associated virus |
|
|
| Mint vein banding-associated virus-US | [AY548173*] | (MVBaV-US) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequence does not comprise the complete genome.
List of other related viruses which may be members of the family Closteroviridae but have not been approved as species
| Grapevine leafroll-associated virus 7 | [EF093187*] | (GLRaV-7) |
| Little cherry virus 1 | [Y10237=NC_001836] | (LChV-1) |
| Olive leaf yellowing-associated virus | [AJ440010*] | (OLYaV) |
* Sequences do not comprise the complete genome.
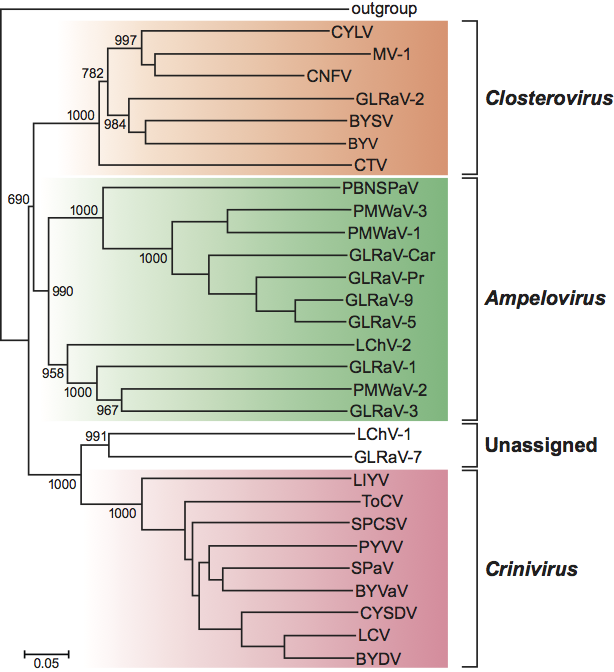
Phylogenetic relationships within the family
Phylogenetic relationships within the family are depicted in Figure 6.
Similarity with other taxa
Virions of some of the genera of the families Alphaflexiviridae (Allexivirus) and Betaflexiviridae (Capillovirus, Trichovirus, Vitivirus, Citrivirus and Foveavirus) have the same particle morphology as those of the family Closteroviridae. However, the sequence of the CP of members of this family has little similarity with that of CPs of viruses in the above genera, and major differences exist in genome size and organization, and in strategy of expression. Replication-associated proteins (RdRp, Mtr and Hel) contain signature sequences homologous to those of other taxa of the “alpha-like” supergroup of ssRNA viruses, the closest affinity being with the families Bromoviridae and Virgaviridae. The replication strategy, based on polyprotein processing, translational frameshifting and multiple sgRNA generation, closely resembles that of viruses in the families Coronaviridae and Arteriviridae. However, unlike closteroviruses, the RdRp of coronaviruses and arteriviruses belong to the “picorna-like” supergroup of polymerases. Hence, the transcriptional strategy of members of the family Closteroviridae follows the mechanism of other “alpha-like” viruses, and is dissimilar from the discontinuous, leader-primed transcription of coronaviruses and arteriviruses.
Derivation of names
Ampelo: from Greek ampelos, “grapevine”, the host of the type species of the genus.
Clostero: from Greek kloster, “spindle, thread”.
Crini: from Latin crinis, “hair”, from the appearance of the very long thread-like particles.
Further reading
Agranovsky, A.A., Lesemann, D.E., Maiss, E., Hull, R. and Atabekov, J.G. (1995). Rattlesnake structure of a filamentous plant RNA virus built of two capsid proteins. Proc. Natl. Acad. Sci. USA, 92: 2470-2473.
Alzhanova, D.V., Prokhnevsky, A.I., Peremyslow, V.V. and Dolja, V.V. (2007). Virion tails of Beet yellows virus: coordinated assembly by three structural proteins. Virology, 39, 220-226.
Bar-Joseph, M., Garnsey, S.M. and Gonsalves, D. (1979). The closteroviruses: a distinct group of elongated plant viruses Adv. Virus Res., 25: 93-168.
Boyko, V.P. , Karasev, A.A., Agranovsky, A.A., Koonin, E.V. and Dolja, V.V. (1992). Coat protein gene duplication in a filamentous RNA virus of plants. Proc. Natl. Acad. Sci. USA,89: 9156-9160.
Coffin, R.S. and Coutts, R.H.A (1993). The closteroviruses, capilloviruses and other similar viruses: a short review. J. Gen. Virol., 74, 1475-1483.
Duffus, J.E. (1971). Role of weeds in the incidence of virus diseases. Annu. Rev. Phytopathol., 9, 319-340.
Duffus, J.E. (1973). The yellowing virus diseases of beet. Adv. Virus Res., 18, 347-386.
Faoro, F. (1997). Cytopathology of closteroviruses and trichoviruses infecting grapevines. In: Filamentous Viruses of Woody Plants (P.L. Monette, ed), pp 29-47. Research Signpost, Trivandrum.
Klaassen, V.A., Boeshore, M.L., Koonin, E.V. Tian, T. and Falk, B.W. (1995). Genome structure and phylogenetic analysis of lettuce infectious yellows virus, a whitefly-transmitted, bipartite closterovirus. Virology, 208, 99-110.
Lesemann, D.E. (1988). Cytopathology. In: The Plant Viruses. Filamentous Plant Viruses, Vol. 4, (R.G. Milne, ed), pp 179-235. Plenum Press, New York.
Liu, H.Y., Wisler, C.G. and Duffus, J.E. (2000). Particle length of whitefly-transmitted crinviruses. Plant Dis., 84, 803-805.
Medina, V., Rodrigo, G., Tian T., Juarez, M., Dolja, V.V., Achon M. and Falk, B.W. (2003). Comparative cytopathology of Crinivirus infections in different plant hosts. Ann. Appl. Biol., 143, 99-110.
Milne, R.G. (1988). The economic impact of filamentous plant viruses. In: The Plant Viruses. Filamentous Plant Viruses, Vol. 4 (R.G. Milne, ed), pp. 333-407. Plenum Press, New York.
Salem, N.M., Che, A.Y.S., Tzanetakis, I.E., Mongkolsiriwattana, C. and Ng, C.K. (2009). Further complexity of the genus Crinivirus revealed by the complete genome sequence of Lettuce chlorosis virus (LCV) and the similar temporal accumulation of LCV genomic RNAs 1 and 2. Virology, 390, 45-55.
Contributed by
Martelli, G.P., Agranovsky, A.A., Bar-Joseph, M., Boscia, D., Candresse, T., Coutts, R.H.A., Dolja, V.V., Hu, J.S., Jelkmann, W., Karasev, A.V., Martin, R.R., Minafra, A., Namba, S. and Vetten, H.J.
Figures
Figure 1 (Top) Negative contrast electron micrograph of virions of citrus tristeza virus (CTV) (genus Closterovirus). Particles of all members of the genera Ampelovirus and Crinivirus have a similar morphology. The bar represents 100 nm. (Courtesy of R.G. Milne.) (Bottom) Beet yellows virus (BYV) particles showing a decorated extremity (arrow heads) following exposure to an antiserum to the N-terminal peptide expressed from the minor CP gene. The bar represents 100 nm.
(Courtsey of D.E. Lesemann.)
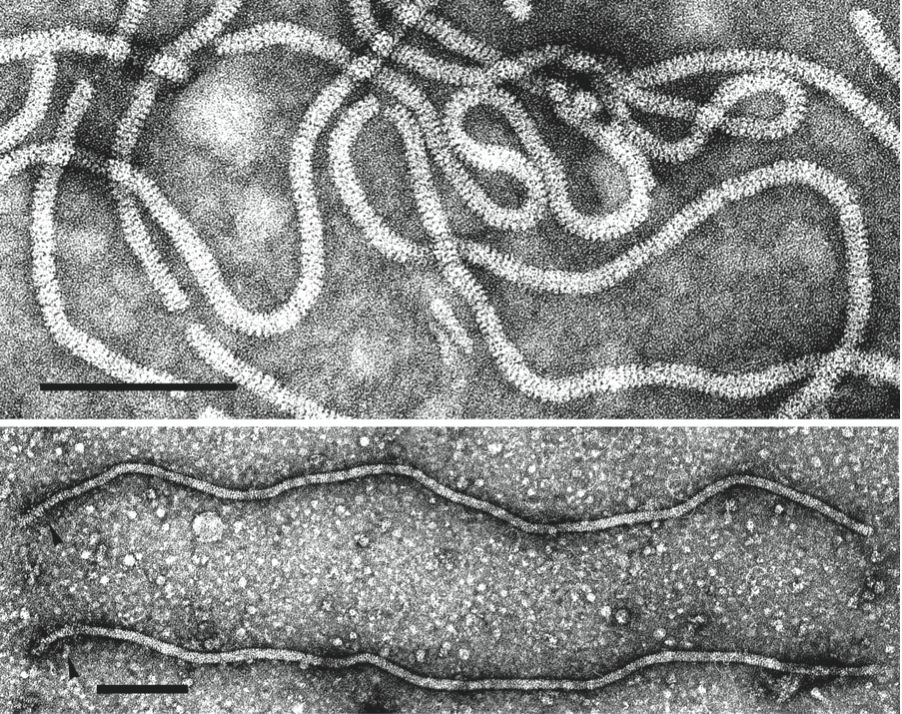
Figure 2 Genome organization and strategy of replication of beet yellows virus (BYV) showing the relative position of the ORFs, their expression products, and the 3 nested set of sgRNAs. L-Pro, leader proteinase (66K); Mtr, methyltransferase (63K); Hel, helicase (100K); RdRp, RNA polymerase; HSP70h; ~60 kDa protein; CP, coat protein; CPm, minor capsid protein. The five boxes under cell-to-cell movement represent the five gene block conserved among most closteroviruses.
(From Dolja, 2003; with permission.)
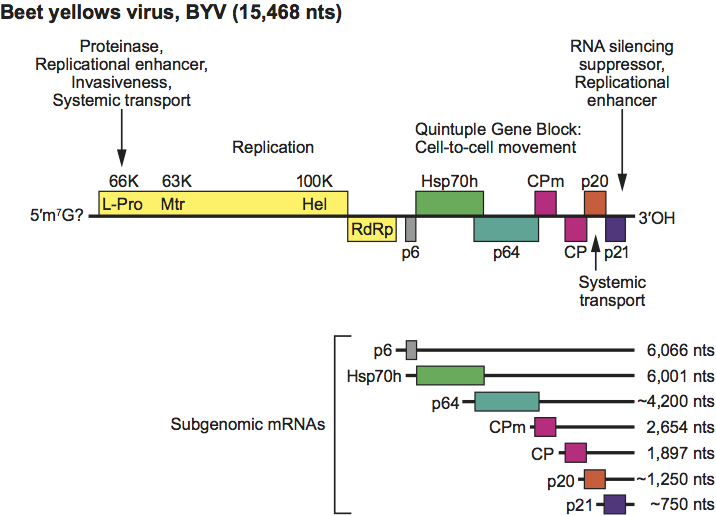
Figure 3 Organization and expression strategy of genomes characteristic of citrus tristeza virus showing the relative position of the ORFs and their expression products. +1 FS the putative ribosomal frameshift. The solid lines at the bottom of the figure define the two genomic regions expressed through proteolytic processing of the polyprotein precursor(s) and through the formation of a nested set of 3 co-terminal sgRNAs. P-Pro, papain-like protease; Mt, methytransferase; Hel, helicase; RdRp, RNA polymerase; HSP70h, ~60 kDa protein; CPm, minor capsid protein; CP, capsid protein.
(From Karasev and Hilf, 1997; with permission.)
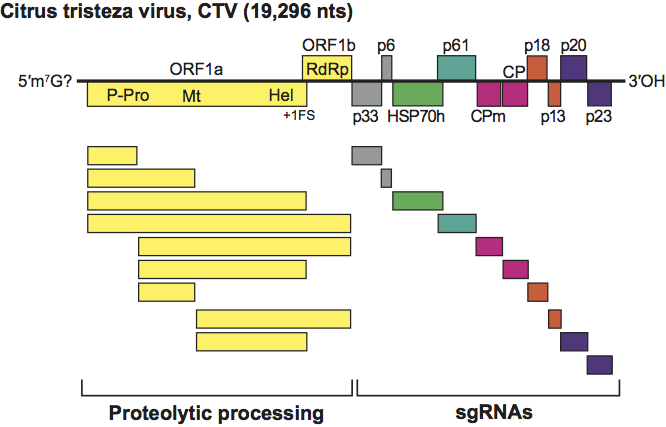
Figure 4 Genome organization of grapevine leafroll-associated virus 3, showing the relative position of the ORFs and their expression products. Pro, papain-like protease; Mtr, methytransferase; Hel, helicase; RdRp, RNA polymerase; HSP70h; ~60 kDa protein; CPm, minor capsid protein; CP, capsid protein.
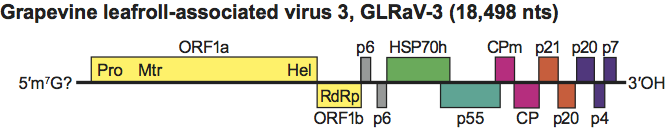
Figure 5 Genome organization of lettuce infectious yellows virus showing the relative position of the ORFs and their expression products. P-Pro, papain-like protease; Mtr, methyltranferase; Hel, helicase; RdRp, RNA polymerase; HSP70h; ~60 kDa protein; CP capsid protein; CPm, minor capsid protein.
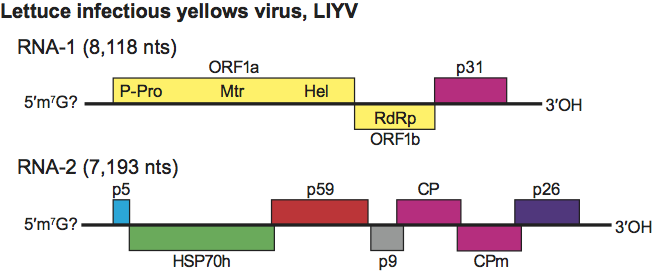
Figure 6 Phylogenetic tree showing the relationships between the species and genera of the family Closteroviridae based on the sequence of the HSP70h gene. The neighbor-joining tree was produced and bootstrapped using CLUSTAL W. Branch lengths are proportional to sequence distances. All abbreviations and accession numbers can be found in the List of Species in the description. GLRaV-7 and LChV-1 are unassigned viruses in the family.
(Courtesy of P. Saldarelli.)