Genus: Cilevirus
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Type species: Citrus leprosis virus C
Distinguishing features
This genus contains mite-transmitted viruses with bacilliform particles and a bipartite ssRNA genome that is polyadenylated.
Virion properties
Morphology
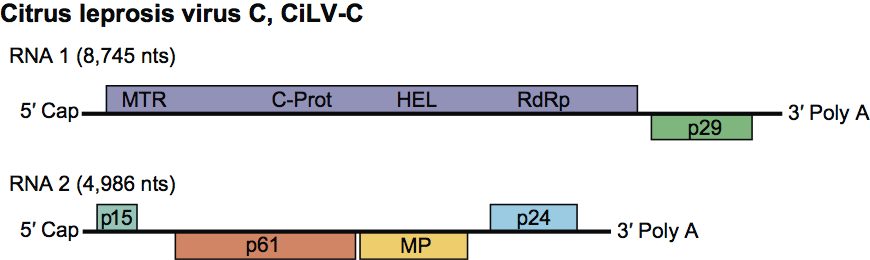
Virions are bacilliform, about 120–130×50–55 nm and are not enveloped (Figure 1). In ultra-thin sections of infected citrus plants, they can be seen in enclaves of the endoplasmic reticulum of mesophyll and vascular parenchyma (Figure 2).
Physicochemical and physical properties
Virions have an in vitro thermal inactivation point between 55 and 60 oC. They remain viable for 6 days at 4 oC and 3 days at room temperature. Dilution end point is 10−3. Infectivity is retained for about 45 months in dried leaves. Virions of Citrus leprosis virus C (CiLV-C) can be isolated 24 h after inoculation from inoculated leaves of Chenopodium quinoa with very low titer of infection.
Nucleic acid
Virions contain two molecules of linear, positive sense, ssRNA of about 8745 nt (RNA 1) and 4986 nt (RNA 2). The RNA molecules are polyadenylated in the 3′-termini, and contain a “cap” structure in the 5′-termini.
Proteins
See section on genome organization and replication.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genome of CiLV-C is bipartite. ORF1 of RNA1 (frame +1) encodes a large polypeptide involved in viral replication. This has 2512aa (286 kDa) and contains conserved domains for methyltransferase (MTR), cysteine protease (C-Prot), helicase (Hel) and RNA dependent RNA polymerase (RdRp). ORF2 of RNA1 (frame +3) encodes a putative protein of 263aa (29 kDa) with no conserved domains but a very low similarity to the capsid proteins of members of the genus Alphavirus. This fact, associated with serological data obtained using antiserum against recombinant P29 suggest that the protein may be involved in virion encapsidation. This ORF is followed by a 3′ NTR of 228 nt and a poly (A) tail of undetermined length (Figure 3). RNA2 has four ORFs. ORF1 (frame +1) encodes a putative protein with 130aa (15 kDa). ORF2 (frame +3) encodes the largest putative protein of this RNA (537aa, 61 kDa). ORF3 (frame +3) codes for a putative protein of 32 kDa (297 aa) with conserved domains characteristic of a viral movement protein (MP). ORF4 (frame +1) codes for a putative protein with 214aa (24 kDa). None of the putative proteins encoded by ORFs 1, 2 and 4 of RNA2 have conserved domains or similarity with known sequences in databases. ORF4 of RNA2 is followed by a 3′ NTR of 232 nt and a poly (A) tail of undetermined length (Figure 3). The conserved sequence GAUAAAUCU was found at the 5′-termini of both RNAs 1 and 2 of CiLV-C. The virus has four subgenomic RNAs (sgRNA). The first one (sgRNA1, 796 nt) corresponds to nucleotide sequences of p29 of RNA1, while the other three correspond to sequences of RNA2 (sgRNA2 of 3 kb corresponds to regions within p61, p32 and p24, sgRNA3 of 1.5 kb transcribes regions of p32 and p24, and sgRNA4 of 0.6 kb corresponds to p24 sequence). Infected citrus leaves contain dsRNAs corresponding to double stranded forms of RNA1 and RNA2 as well as subgenomic RNAs.
Antigenic properties
It has not been possible to purify the virion using standard procedures. In heterologous systems using Eschericha coli, P29 was highly immunogenic, yielding a high quality antiserum. All of the other proteins tested appear less immunogenic.
Biological properties
Transmission occurs by all of the active stages of the mite vector Brevipalpus sp. (Acari: Tenuipalpidae): larvae, nymphs and adults. The virus circulates but, differently from what has been reported earlier, it does not seem to propagate in the vector. The virus was initially considered to have a very narrow host range, but in recent years the list of CiLV-C hosts has increased significantly. In nature, CiLV-C infects several species of Citrus. Sweet oranges (Citrus sinensis) are among the most susceptible species, along with C. keraji. Mandarins (C. reticulata, C. reshni, C. deliciosa, C. lycopersiciformis, C. unshiu, C. depressa) and few hybrids exhibit variable levels of resistance, but are symptomatic. The rutaceous plant Swinglea glutinosa has been recently reported as the first natural host of CiLV-C outside the genus Citrus. Chenopodium quinoa, C. amaranticolor and Gomphrena globosa are experimental hosts by mechanical inoculation only, while CiLV-C can also be experimentally transmitted to Solanum violaefolium, Phaseolus vulgaris, Hibiscus rosa-sinensis, Malvaviscus arboreus, Grevilea robusta, Bixa orellana, Glycosmis pentaphylla and Commelina benghalensis by the mite vector. Graft transmission of CiLV-C is possible experimentally but, contrary to what happens to other citrus viruses, it does not occur naturally. The virions remain confined to the localized lesions they induce, and are never spread systemically within the host plants. Typical symptoms are chlorotic or necrotic lesions, often exhibiting ringspot patterns in leaves, fruits and stems. Lesions can be raised in all affected organs but, in fruits, they can also be depressed. Affected leaves and fruits may drop prematurely. Heavy infection can eventually lead to the death of the tree. Typically from the Americas, CiLV-C is present in Paraguay, Argentina, Brazil, Bolivia, Venezuela, Colombia, Panama, Costa Rica, Guatemala, Nicaragua, Honduras, El Salvador and Mexico. Its first report was from the United States, but it has not been found in that country since the 1960s. There are unconfirmed reports of the occurrence of leprosis in Asia and Africa.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Cilevirus
| Citrus leprosis virus C |
|
|
| Citrus leprosis virus C - Cordeiropolis | RNA1 [DQ352194]RNA2 [DQ352195] | (CiLV-C-Cor) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Cilevirus but have not been approved as species
| Ligustrum ringspot virus | [HM164551*], [HM164552*] | (LigRSV) |
| Solanum violaefolium ringspot virus | [DQ 514336*] | (SvRSV) |
| Passion fruit green spot virus | [HM002746*], [HM002747*] | (PFGSV) |
* Sequences do not comprise the complete genome.
Similarity with other taxa
CiLV-C and other possible species of the genus Cilevirus have unusual biological properties such as the inability to spread systemically within the host plants, the Brevipalpus mite transmission and the uncommon bacilliform morphology of the virions. Sequence analyses reveal conserved domains corresponding to the movement protein and replication-associated proteins of members of several positive sense, ssRNA genera such as Furovirus, Bromovirus, Tobravirus and Tobamovirus. However, none of the other four ORFs of CiLV-C or their translated proteins exhibit conserved domains or similarity with sequences available in the main sequence databases.
Derivation of name
Cilevirus: from the genus of the natural host (Citrus) and the name of the disease caused by the type member virus (leprosis).
Further reading
Bastianel et al., 2010 M. Bastianel, V.M. Novelli, E.W. Kitajima, K.S. Kubo, R.B. Bassanezi, M.A. Machado, J. Freitas-Astúa, Citrus leprosis: centennial of an unusual mite virus pathosystem. Plant Dis. 94 (2010) 284–292.
Freitas-Astúa et al., 2008 J. Freitas-Astúa, F. Nicolini, M. Bastianel, E.W. Kitajima, Interações entre o vírus da leprose dos citros e seu vetor Brevipalpus phoenicis. Summa Phythopathol. 34 (2008) 128–130.
Locali-Fabris et al., 2006 E.C. Locali-Fabris, J. Freitas-Astúa, A.A. Souza, M.A. Takita, G. Astúa-Monge, R. Antonioli-Luizon, V. Rodrigues, M.L.P.N. Targon, M.A. Machado, Complete nucleotide sequence, genomic organization and phylogenetic analysis of Citrus leprosis virus cytoplasmic type. J. Gen. Virol. 87 (2006) 2721–2729.
Lovisolo, 2001 O. Lovisolo, Citrus leprosis virus, properties, diagnosis, agro-ecology, and phytosanitary importance. EPPO Bull. 31 (2001) 79–89.
Pascon et al., 2006 R.C. Pascon, J.P. Kitajima, M.C. Breton, L. Assumpção, C. Greggio, A.S. Zanca, V.K. Okura, M.C. Alegria, M.E. Camargo, G.G.C. Silva, C.J. Cardoso, MA. Vallim, S.F. Franco, V.H. Silva, H. Jordão Júnior, F. Oliveira, P.F. Giachetto, F. Ferrari, C.I. Aguilar-Vildoso, F.J.B. Franchiscini, J.M.F. Silva, P. Arruda, J.Á. Ferro, F. Reinach, A.C.R. Silva, The complete nucleotide sequence and genomic organization of Citrus leprosis associated virus, cytoplasmatic type (CiLV-C). Virus Genes. 32 (2006) 289–298.
Contributed by
Locali-Fabris, E.C., Freitas-Astúa, J. and Machado, M.A.
Figures
Figure 1 Electron micrograph of purified particles of an isolate of Citrus leprosis virus C.
(Courtesy Addolorata Colariccio, Instituto Biolgico, SP, Brazil.)
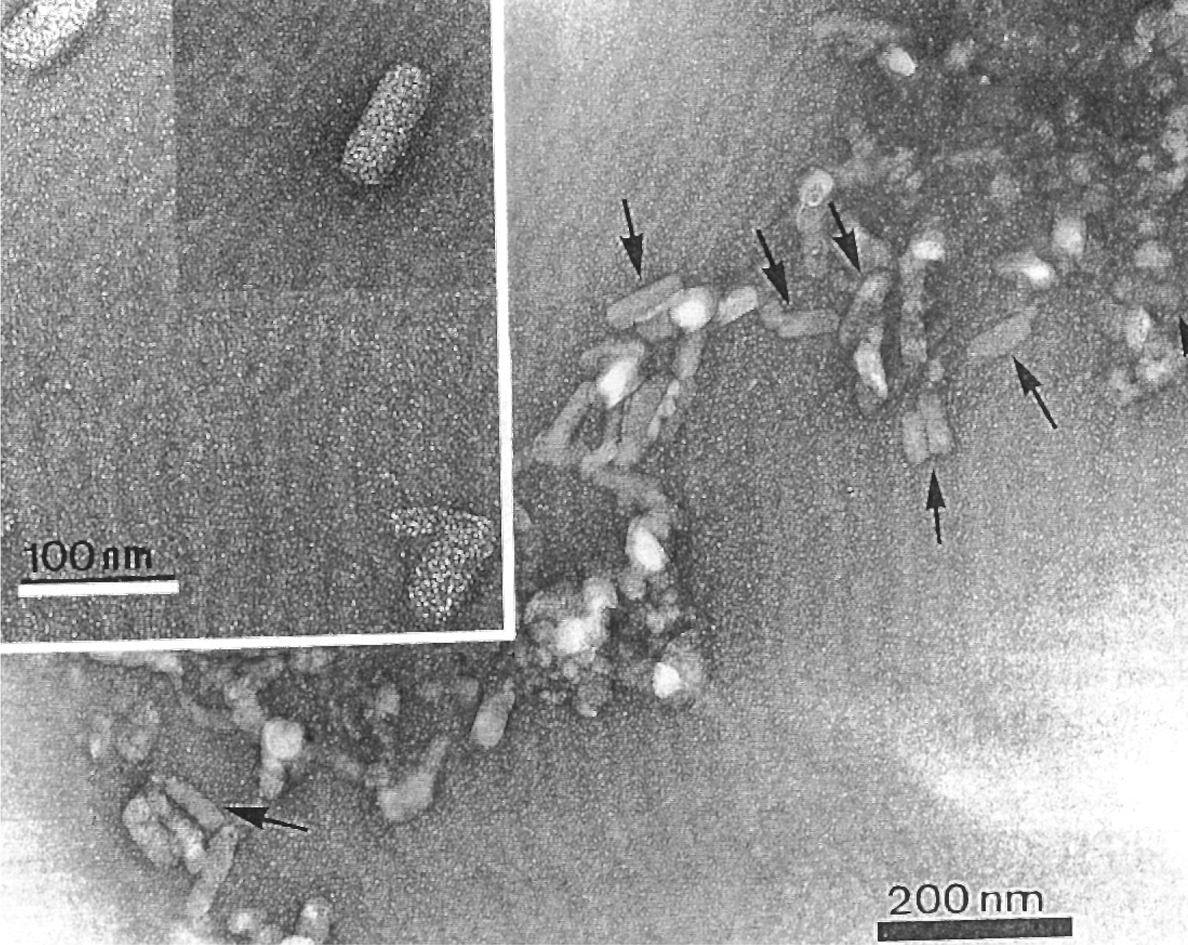
Figure 2 Electron micrographs exhibiting viroplasma (VP) and virions (VI) inside the endoplasmic reticulum in parenchyma cells of Pera sweet orange.
(Courtesy Elliot W. Kitajima, ESALQ/USP, Brazil.)
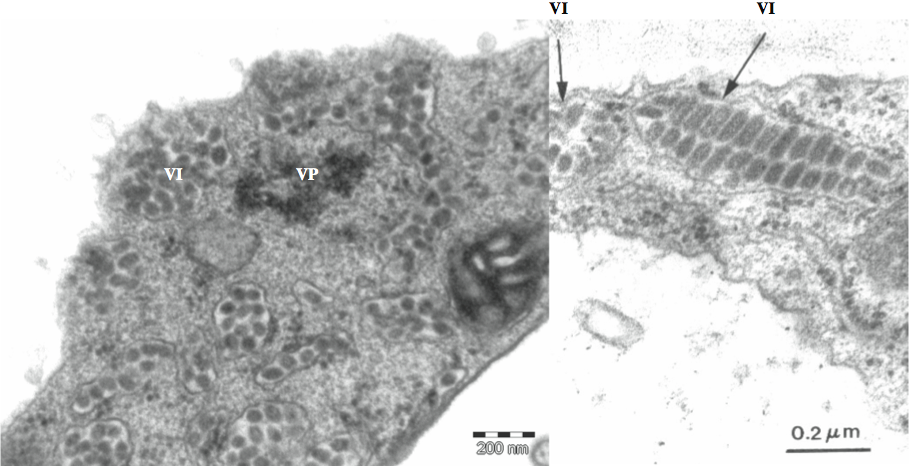
Figure 3 Schematic drawing of genome structure and organization of CiLV-C. Each RNA is presented as a line with the corresponding ORFs indicated by rectangles. ORFs located in different frames are represented above (+1) or below (+3) the lines. Functional domains attributed according to the translated sequences are indicated inside the rectangles. For those translated sequences with unidentified function, the approximate molecular weight is indicated inside the rectangles. MTR=methyltransferase domain; C-Prot=cysteine protease domain; HEL=helicase domain; RdRp=RNA-dependent RNA polymerase domain; MP=movement protein domain.
(Reproduced from Locali-Fabris et al. (2006). J. Gen Virol., 87, 2721-2729; with permission.)