Family: Caliciviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are non-enveloped with icosahedral symmetry. They are 27–40 nm in diameter by negative-stain electron microscopy and 35–40 nm by cryo-electron microscopy (Figure 1). The capsid is composed of 90 dimers of the major structural protein VP1 arranged on a T=3 icosahedral lattice. In noroviruses, the VP1 forms a subunit comprised of a shell and two protruding domains (Figure 1). A characteristic feature of calicivirus capsid architecture is the 32 cup-shaped depressions at each of the icosahedral five-fold and three-fold axes. In negative-stain virus preparations, some cup-shaped depressions appear distinct and well defined, while in others these depressions are less prominent.
Physicochemical and physical properties
Virion Mr is about 15×106. Virion buoyant density is 1.33–1.41 g cm−3 in CsCl and 1.29 g cm−3 in glycerol-potassium tartrate gradients. Virion S20,w is 170–187S. Physicochemical properties have been established for some members of the family. Generally, caliciviruses are stable in the environment and many strains are resistant to inactivation by heat and certain chemicals (ether, chloroform and mild detergents). Enteric caliciviruses are acid-stable.
Nucleic acid
The genome consists of a linear ssRNA molecule of 7.4–8.3 kb. A protein (VPg, 10–15 kDa) is covalently attached to the 5′-terminus of the genomic RNA and the 3′-terminus is polyadenylated (Figure 2). Caliciviruses have conserved nucleotide motifs at the genomic 5′-terminus and at the junction of the coding sequences for the non-structural/structural proteins. A subgenomic RNA (sgRNA) of 2.2–2.4 kb is synthesized intracellularly and is apparently VPg-linked. The gene order for several caliciviruses was determined by transient expression and in vitro translation studies and cleavage mapping yielding at least six mature non-structural proteins, followed by two structural proteins (VP1 and VP2). All the non-structural proteins are encoded by a large polyprotein and expressed in the same order for each genus (Figure 2). In the case of lagoviruses, sapoviruses and neboviruses the open reading encoding the non-structural proteins is in-frame and contiguous with VP1.
Proteins
Virions are predominantly comprised from one major capsid protein, VP1 (58–60 kDa). A second minor structural protein named VP2 (8.5–23 kDa) has been found in association with FCV and RHDV virions, and Norwalk virus-like particles. Some non-structural proteins have sequence and motif homology with those of the family Picornaviridae replicative enzymes and include 2C (NTPase), 3C (cysteine protease) and 3D (RNA-dependent RNA polymerase) domains. The calicivirus VPg (10–15 kDa) is covalently linked to the viral RNA and maps to the region of the calicivirus genome analogous to the 3B region of picornaviruses, but without having an apparent amino acid homology with those of picornaviruses (Figure 2).
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Caliciviruses have single stranded, positive sense genomic RNA (positive-strand RNA) organized into either two or three major ORFs (see genus descriptions). The non-structural proteins are encoded in the 5′-terminus of the genome and the structural proteins in the 3′-terminus (Figure 2). Replication occurs in the cytoplasm and two major positive-strand RNA species are found in infected cells: (1) the genome-sized, positive-strand RNA serves as the template for translation of a large polyprotein that undergoes cleavage by a virus-encoded protease to form the mature non-structural proteins and (2) a subgenomic-sized, positive-strand RNA, co-terminal with the 3′- terminus of the genome is the template for translation of the VP1 as well as the 3′-terminal ORF product VP2. A dsRNA corresponding in size to full-length genomic RNA has been identified in FCV and San Miguel sea lion virus (SMSV)-infected cells, indicating that replication occurs via a negative-strand intermediate.
Antigenic properties
Taking each genus in turn, the prototype virus for the vesiviruses is vesicular exanthema of swine virus (VESV) isolated from pigs and 13 serotypes are recognized by serum neutralization assays. Following the isolation of SMSV, which is the progenitor of VESV, additional vesivirus serotypes were recovered from at least 28 marine and other species. These viruses (now more than 40 serotypes) have been grouped together phylogenetically as the “marine vesiviruses”. FCV represents the second species within the vesiviruses. Although initial studies of FCV indicated there were multiple serotypes, examinations of large collections of viruses and feline antisera obtained from experimental infections in specific pathogen-free cats led to recognition of extensive cross-reactions and a consensus that FCV represents a single serotype, albeit with antigenic variants.
Within the lagoviruses, cross-challenge studies in the natural host and experiments with monoclonal antibodies indicate that RHDV and European brown hare syndrome virus (EBHSV) are antigenically distinct.
Antigenic types have been defined by cross-challenge studies, immune electron microscopy or solid phase immune electron microscopy for non-cultivatable viruses in the genera Norovirus and Sapovirus. By contrast, viruses in the Nebovirus genus appear to fall into a single, closely related antigenic grouping.
For some non-cultivable viruses that are members of the genera Lagovirus, Norovirus and Sapovirus, recombinant virus-like particles (rVLPs) can be produced by expression of VP1 in mammalian, insect and plant systems. These VLPs are highly immunogenic and similar in antigenicity to native virions and they are useful to study the structure and antigenicity of virus particles.
Biological properties
Caliciviruses have been isolated from a broad range of vertebrates. Except for some vesiviruses, individual caliciviruses generally exhibit a natural host restriction and this has presented a considerable problem with studying specific viruses for which there is no productive cell culture system, e.g. human noroviruses. However, at least one example virus from each genus can now be cultivated except for the neboviruses.
The marine vesiviruses have both a broad host range and a wide cell tropism. The diversity in host species of the marine vesiviruses was not initially recognized. For example, the VESVs isolated from swine from 1932 through 1956 were considered swine viruses and those subsequently isolated from ocean or other sources were either designated SMSVs or named after the original host species from which they were first isolated (e.g. walrus, cetacean, reptile). All these marine vesiviruses including those isolated from bystander host species are closely related genetically and generally lack host specificity.
Transmission of caliciviruses is via direct contact with an infected host or indirectly via contact with faecal material, vomitus or respiratory secretions, contaminated food, water and fomites. In general, no biologic vectors appear to be involved in transmission, however, mechanical, arthropod vector transmission of RHDV and VESV has been described.
Caliciviruses are associated with a number of disease syndromes. Marine vesiviruses (e.g. VESV) produced clinical signs in swine that were indistinguishable from foot-and-mouth disease, and include vesicles in the mouth, tongue, lips, snout and feet at the coronary band and between the digits. In addition, the virus may cause encephalitis, myocarditis, fever, diarrhoea, abortion, hepatitis, pneumonia and hemorrhage and failure of recovered animals to thrive. FCV in cats is predominantly associated with oral ulceration and rhinitis, and causes persistent infections. Recently, a more severe disease termed virulent-systemic feline calicivirus disease has been recognized, associated with epithelial cytolysis and systemic vascular compromise in susceptible cats, leading to severe oedema and high mortality.
In the genus Lagovirus, RHDV is associated with a generalized viraemic infection in which there is massive liver necrosis that triggers a disseminated intravascular coagulation and rapid death in rabbits greater than three months of age. Non-virulent viruses related to RHDV have been described. EBHSV is similar to RHDV but appears to be less virulent.
Human caliciviruses in the genera Norovirus and Sapovirus induce a generally self-limiting gastroenteritis. Vomiting is a consistent and prominent symptom; other symptoms may include nausea, diarrhoea, abdominal cramping, fever and malaise. Prolonged disease is seen in immunocompromised individuals. Neboviruses cause diarrhoea and mild upper intestinal lesions in calves.
Genus Vesivirus
Type species Vesicular exanthema of swine virus
Distinguishing features
The species in this genus form a distinct clade within the family (Figure 3). The genome is organized into three major ORFs. ORF1 encodes the non-structural polyproteins. ORF2 encodes the major structural capsid protein that is translated as a larger precursor protein before cleavage into the mature VP1, a feature that appears unique to this genus. ORF1 and ORF2 of the viruses in this genus are separated by either 2 nt (GC for FCV strains) or 5 nt (CCACT/C for marine vesiviruses). A third ORF (ORF3) encodes VP2 a small, basic protein and overlaps by one nt with ORF2 in a −1 frameshift. The ORF3 product has been detected in FCV-infected cells and is essential for infectious virus production.
Biological properties
Most members of this genus can be readily propagated in cell culture. FCV grows most efficiently in cells of feline origin; in vivo, the primary site of replication is the upper respiratory tract. Primary isolation of most marine vesiviruses is possible in African green monkey kidney or porcine kidney cells. When tested approximately half of marine vesiviruses have produced skin vesicles in swine, horses and sometimes cattle and vesicles have occurred naturally in dogs, pinnipeds, cetaceans and primates (including man).
Species demarcation criteria in the genus
Members of a Vesivirus species share a major phylogenetic branch within the genus and a common genome organization, however, demarcation into two species is on the basis of phylogenetic differences, subtle variation in genome structure and host range.
The virus VESV A48 is the prototype virus of the genus Vesivirus. This virus and those isolated from marine species are phylogenetically grouped together and colloquially referred to as “marine vesiviruses”. Feline calicivirus (FCV) is also recognized as a separate species within the genus. A collection of representative vesivirus isolates from which some sequences have been used to generate Figure 3 are shown in the table below.
List of species in the genus Vesivirus
| Feline calicivirus |
|
| |
| Feline calicivirus [F9] | [M86379] | (FCV) | |
| Vesicular exanthema of swine virus |
|
| |
| Bovine calicivirus Bos-1 [217T] | [U18470; U18741; U76875; U76876] | (BCV-Bos-1) | |
| Bovine calicivirus Bos-2 [unnamed] | [AY069947] | (BCV-Bos-2) | |
| California sea lion vesivirus [02012181] | [DQ666635] | (CSLV) | |
| Cetacean calicivirus Tur-1 [78-0041] | [U18471; U52091 ] | (CCV-Tur-1) | |
| Human calicivirus [McA11] |
| (HuCV) | |
| Primate calicivirus Pan-1 [78-0029] | [AF091736] | (PCV-Pan-1) | |
| Rabbit calicivirus [4] | [AJ866991] | (RaCV) | |
| Reptile calicivirus Cro-1 [78-0002] | [U52092; AY772542] | (RCV-Cro-1) | |
| San Miguel sea lion virus 1 [1MR] | [U15301] | (SMSV-1) | |
| San Miguel sea lion virus 2 [2MR] | [DQ666629] | (SMSV-2) | |
| San Miguel sea lion virus 4 [15FT] | [M87482] | (SMSV-4) | |
| San Miguel sea lion virus 5 [205-73] | [U18477; U18731; U52093; U76883; U76884; DQ300285] | (SMSV-5) | |
| San Miguel sea lion virus 6 [439] | [U18478; U18732; U76885; AJ131387; U76886] | (SMSV-6) | |
| San Miguel sea lion virus 7 [Gn-21] | [U18479; U18733; U76887; U76888] | (SMSV-7) | |
| San Miguel sea lion virus 8 [274] |
| (SMSV-8) | |
| San Miguel sea lion virus 9 [427] | [U52090] | (SMSV-9) | |
| San Miguel sea lion virus 10 [V-31-77] | [DQ666631] | (SMSV-10) | |
| San Miguel sea lion virus 11 [804T] |
| (SMSV-11) | |
| San Miguel sea lion virus 12 [2615T] |
| (SMSV-12) | |
| San Miguel sea lion virus 13 [CSL-461] | [DQ666636] | (SMSV-13) | |
| San Miguel sea lion virus 14 [A1280] | [U18475; U18735; U76879; U76880] | (SMSV-14) | |
| San Miguel sea lion virus 15 [220R] | [U52088] | (SMSV-15) | |
| San Miguel sea lion virus 16 [Zc-698] | [U52089] | (SMSV-16) | |
| San Miguel sea lion virus 17 [Zc-91PG] | [U52094; U52005] | (SMSV-17) | |
| Skunk calicivirus [4-1L] | [U14667; U14668] | (SCV) | |
| Steller sea lion vesivirus V810 [SSL2002-379SE] | [EF193004] | (SSLV-V810) | |
| Steller sea lion vesivirus V1415 [SSLV-V1415] | [EF195384] | (SSLV-V1415) | |
| Vesicular exanthema of swine virus 1934B [B1-34] | [U18480; U18736] | (VESV-1934B) | |
| Vesicular exanthema of swine virus A48 [Fontana/48] | [U76874] | (VESV-A48) | |
| Vesicular exanthema of swine virus B51 [Davis/51] | [DQ666632] | (VESV-B51) | |
| Vesicular exanthema of swine virus C52 [San Francisco/52] | [U18482; U18738; DQ666633] | (VESV-C52) | |
| Vesicular exanthema of swine virus D53 [Riverside/53] |
| (VESV-D53) | |
| Vesicular exanthema of swine virus E54 [Warm Springs/54] | [U18483; U18739] | (VESV-E54) | |
| Vesicular exanthema of swine virus F55 [FS 330] |
| (VESV-F55) | |
| Vesicular exanthema of swine virus G55 [FS 332] | [DQ666637] | (VESV-G55) | |
| Vesicular exanthema of swine virus H54 [FS 315] |
| (VESV-H54) | |
| Vesicular exanthema of swine virus I55 [GF 1] | [U18484; U18740] | (VESV-I55) | |
| Vesicular exanthema of swine virus J56 [Sec. 3-56] |
| (VESV-J56) | |
| Vesicular exanthema of swine virus K54 [Sec. 3-54] | [DQ666634] | (VESV-K54) | |
| Walrus calicivirus [7420] | [AF321298] | (WCV) | |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Vesivirus but have not been approved as species
| Canine calicivirus [No. 48] | [AB070225] | (CaCV) |
| 2117 CHOcalicivirus | [AY343325] | (2117) |
| Mink calicivirus [MV-20] | [AF338407] | (MCV) |
Genus Lagovirus
Type species Rabbit hemorrhagic disease virus
Distinguishing features
The strains in this genus form a distinct clade within the family. The genome is organized into two major ORFs. ORF1 encodes the non-structural polyprotein, with the major structural protein gene (VP1) in frame with the non-structural polyprotein coding sequence. The translated region of ORF2 overlaps ORF1 by 5 nt in the RHDV genome and 5 nt in the EBHSV genomes. The ORF2 encodes a small protein (VP2) of unknown function that has been identified as a minor structural component in the RHDV virion.
Biological properties
These viruses have characteristically been associated with infection in rabbits and hares (lagomorphs), and can cause epidemics with high mortality in these animals. There is no productive continuous cell culture system for lagoviruses but recently infection of a rabbit kidney cell line with RHDV has been reported.
Species demarcation criteria in the genus
Members of a Lagovirus species, either RHDV or EBHSV, share a major phylogenetic branch within the genus, common genome organization, natural host range and cross-protection antigens. Examples of lagoviruses are shown in Table 3.
List of species in the genus Lagovirus
| European brown hare syndrome virus |
|
|
| EBHSV [FRG] | [U09199] | (EBHSV-FRG) |
| EBHSV [GD] | [Z69620] | (EBHSV-GD) |
| Rabbit hemorrhagic disease virus |
|
|
| RHDV [FRG/89] | [M67473] | (RHDV-FRG) |
| RCV [ITL/95] | [X96868] | (RHDV-RCV) |
| RCV-A1 [MIC-07/2007/AU] | [EU871528] | (RHDV-RCV-A1) |
| RHDV [Ashington/1998/UK] | [EF558587] | (RHDV-Ash) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Lagovirus but have not been approved as species
None reported.
Genus Norovirus
Type species Norwalk virus
Distinguishing features
The strains in this genus form a distinct clade within the family. The full-length genome sequence is available for many strains (100+). The first complete genomes to be sequenced were for Norwalk virus, Southampton virus and Lordsdale virus and showed that the genome is organized into three major ORFs. ORF1 encodes the non-structural polyprotein. ORF2 encodes the major structural capsid protein (VP1) and overlaps by 14 nt with ORF1 in the Norwalk and Southampton virus strains and by 17 nt in the Lordsdale virus strain, resulting in a −2 frameshift of ORF2 in all three viruses. ORF3 overlaps by one nt with ORF2 in a −1 frameshift and encodes a small virion-associated protein (VP2). Detailed phylogenetic analysis indicated there are at least five “genogroups” (labelled GI to GV). Examples of the key viruses that form the genogroups are shown in Figure 3 and are listed below. Genogroup I (GI) is made up of eight genotypes, GII contains 19 genotypes, GIII and GIV each have two genotypes and GV has only one.
Biological properties
Noroviruses have been detected in several mammalian species. Human noroviruses belong to GI, GII and GIV. Porcine (GII), bovine and ovine noroviruses (GIII) have been described and murine noroviruses currently constitute GV. However, serological evidence suggests other rodent noroviruses exist and thus viruses in GV are likely not to be exclusively murine in origin. At present only murine noroviruses can be grown in cell culture. Other mammalian species can be experimentally infected with human noroviruses. Furthermore, noroviruses closely related to human noroviruses have been identified from other mammalian species, but it is not clear whether these are the natural hosts or bystanders that have become naturally infected. Recombination between viruses in the same genotype and between genogroups occurs.
Species demarcation criteria in the genus
Not applicable as only one species is currently recognized. However, the range of host species and the nature and extent of diversity of the noroviruses will need further characterization to delineate species criteria.
List of species in the genus Norovirus
There is only one species, Norwalk virus, however, representatives of each genogroup are shown below:
| Norwalk virus |
|
|
| GI [Hu/Norwalk virus 8FiiA/68/US] | [M87661] | (GI-NV) |
| GI [Hu/Southampton/1991/UK] | [L07418] | (GI-SHV) |
| GII [Hu/Hawaii virus/1971/US] | [U07611] | (GII-HV) |
| GII [Hu/Lordsdale/1993/UK] | [X86557] | (GII-LDV) |
| GII [Po/Sw918/1997/JP] | [AB074893] | (GII-Sw918) |
| GIII [Bo/Jena/1980/GE] | [AJ011099] | (GIII-Jena) |
| GIII [Bo/Newbury2/1976/UK] | [AF097917] | (GIII-Newbury2) |
| GIV [Hu/Alphatron/98-2/1998/NL] | [AF195847] | (GIV-Alphatron) |
| GIV [Hu/Fort Lauderdale/560/1998/US] | [AF414426] | (GIV-FtLauderdale) |
| GV [Mu/Murine norovirus 1/2002/US] | [AY228235] | (GV-MNV1) |
| GVI* [Hu/Chiba/040502/2004/JP] | [AJ844470] | (GVI-Chiba) |
| GVI* [Ca/Bari/91/2007/IT] | [FJ875027] | (GVI-Bari) |
| GVI* [Ca/C33/Viseu/2007/PT] | [GQ443611] | (GVI-Viseu) |
Species names are in italic script. Names of genogroups (G) followed by representative virus designations are in roman script. Host abbreviations are: Hu, human: Bo, bovine. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Norovirus but have not been approved as species
None reported.
Genus Sapovirus
Type species Sapporo virus
Distinguishing features
The viruses in this genus form a distinct clade within the family. The first full-length genomic sequence available was for the human virus Manchester virus and subsequently for a range of viruses including the “porcine enteric calicivirus” (PEC strain Cowden). The Manchester virus genome is organized into three ORFs, two with known function. ORF1 encodes the non-structural polyprotein together with the major structural capsid protein gene (VP1) in frame with the non-structural polyprotein coding sequence. In PEC, ORF2 overlaps ORF1 in a −1 frameshift and encodes a predicted small protein likely the equivalent of VP2 in noroviruses. ORF3 begins 11 nt downstream from the predicted start codon of the VP1 in a +1 frameshift and encodes a predicted protein of ~160 aa. In some strains of this genus, and in the PEC Cowden strain, ORF3 is absent. Viruses in the genus are highly heterogeneous and can be divided on the basis of phylogenetic analysis of VP1 into five genogroups; GI, GII, GIV and GV contain human sapoviruses whereas porcine sapoviruses are found only in GIII (Figure 3). Each sapovirus genogroup can be subdivided into numerous genotypes. Examples of sapoviruses representing the genogroups are shown below.
Biological properties
None of the human sapoviruses can be propagated in cell culture. However, PEC can be grown in porcine kidney cells as long as the media is supplemented with bile salts to induce intracellular signals to permit virus replication. Sapovirus infection most commonly occurs in children under the age of 5 years and is especially characterized by outbreaks in closed settings. However, recent molecular epidemiological studies have suggested that the number of outbreaks in which individuals of all age groups are infected may be increasing. Expression of VP1 in recombinant systems usually results in the production of VLPs. Antiserum raised against these VLPs and cross-reactivity studies using these VLPs and antiserum suggests that the antigenic differences amongst sapoviruses mirror the phylogenetic differences observed by genogrouping. Inter-genogroup and intra-genogroup recombination has been documented.
List of species in the genus Sapovirus
The prototype strain for the species is the Sapporo virus, however, there is only a partial genome sequence available for this virus. Other representative viruses (one for each genogroup) are shown below.
| Sapporo virus |
|
|
| GI [Hu/Sapporo/1982/JP] | [U65427] | (GI-SV) |
| GI [Hu/Manchester/1993/UK] | [X86560] | (GI-Man) |
| GII [Hu/London 29845/1992/UK] | [U95645] | (GII-Lon) |
| GII [Hu/Bristol/1998/UK] | [AJ249939] | (GII-Bristol) |
| GIII [Po/PEC Cowden] | [AF182760] | (GIII-Cowden) |
| GIII [Po/sav1/2008/CHN] | [FJ387164] | (GIII-SAV1) |
| GIV [Hou7-1181/1990/US] | [AF435814] | (GIV-Houston) |
| GIV [Hu/Angelholm/SW278/2004/SE] | [DQ125333] | (GIV-Angelholm) |
| GV [Hu/Arg39/1995/ARG] | [AY289803] | (GV-Arg39) |
| GV [NongKhai-24/THA] | [AY646856] | (GV-NongKhai24) |
| GVI* [Po/OH-JJ681/2000/US] | [AY974192] | (GVI-OH-JJ681) |
| GVI* [Po/OH-LL26/2002/US] | [AY974195] | (GVI-OH-LL26) |
| GVII* [Mk/MEC/1/1999/US] | [AF338404] | (GVII-MEC/1/1999) |
| [Po/43/06-18p3/06/ITA†] | [EU221477] | (43/06-18p3) |
| [Po/2053P4/Brazil†] | [DQ359100] | (2053P4) |
| [Po/F19-10/CAN†] | [FJ498786] | (F19-10) |
Species names are in italic script. Names of genogroups (G) followed by representative virus designations are in roman script. Host abbreviations are: Hu, human; Po, swine. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed. The genome sequence for Sapporo virus is only partial, therefore the first complete sapovirus genome sequence is for Manchester virus.
List of other related viruses which may be members of the genus Sapovirus but have not been approved as species
None reported.
Genus Nebovirus
Type species Newbury-1 virus
Distinguishing features
The viruses in this genus have been detected only in calves thus far and they form a distinct clade within the family. The complete genome sequence is available for several strains including Newbury “agent”-1, the prototype virus from the UK, and BEC-NB (Nebraska) virus described in the USA. The nebovirus genome is organized into two major ORFs. ORF1 encodes the non-structural polyprotein, with the major structural capsid protein (VP1) gene in frame with the non-structural polyprotein. VP2 is frameshifted +2 and begins 2 nts after the termination of ORF1.
Biological properties
Neboviruses have been known since the 1980s but constitute the most recently defined calicivirus genus. They cause endemic diarrhoeal disease in calves and the severity of disease in experimentally infected calves is considered equal or worse than that caused by genogroup 3 bovine noroviruses. Neboviruses cannot be grown in cell culture and therefore information on their biological properties is limited. There appear to be two phylogenetically distinct polymerase types but the VP1 sequence is conserved between the prototype viruses, Newbury agent −1 and BEC NB virus. This suggests that, like other enteric caliciviruses, neboviruses may also undergo recombination.
Species demarcation criteria in the genus
Not applicable as only one species is currently recognized. Additional characterization of the viruses in this genus will be required in order to delineate species criteria.
List of species in the genus Nebovirus
| Newbury-1 virus |
|
|
| Newbury-1 virus UK-1976 | [DQ013304] | (Newbury1) |
| Bovine calicivirus CV23-OH | [AY082890] | (CV23-OH) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Nebovirus but have not been approved as species
None reported.
List of unassigned species in the family Caliciviridae
None reported.
List of other related viruses which may be members of the family Caliciviridae but have not been approved as species
In 2008, a positive-strand RNA virus (Tulane virus) was isolated from stool samples taken from rhesus macaques held in captivity. The genome sequence and organisation (three ORFs) of this virus show the characteristic structure of caliciviruses (as defined in Figure 2) although with 6714 nt it is some 600 nt shorter than previously described calicivirus genomes. Whilst Tulane virions share similar morphology and density (1.7 g cm−3) to other caliciviruses, phylogenetic analysis of the Tulane virus genome placed it on a deeply rooted branch separate from the other calicivirus genera. Although no disease entity has been associated with the Tulane virus, it appears to be an enteric virus and is cultivatable in a continuous macaque kidney cell line. The genus name Recovirus (for rhesus enteric calicivirus) has been proposed within the scientific literature with Tulane virus as the prototype.
In 2009 a report described a novel calicivirus isolated from pigs in Quebec, Canada. Genomic analysis revealed a positive sense genome of 6409 nt encoding two major ORFs. Phylogenetic analysis showed that these viruses form a unique cluster with a common root with the noroviruses and the Tulane virus. The genus name Valovirus was proposed with the St-Valérien virus as the prototype.
In October 2010 the complete genome sequence of a chicken calicivirus was submitted to the public databases. Analysis of this sequence suggests that it may form a novel calicivirus species and genus.
| Tulane virus | [EU391643] | (TulaV) |
| St-Valérien virus | [FJ355928] | (ValoV) |
| Chicken calicivirus | [HQ010042] | (ChCV) |
Phylogenetic relationships within the family
There are five recognized genera within the family Caliciviridae. The phylogenetic relationships amongst them are summarized in Figure 3.
Similarity with other taxa
Caliciviruses have some properties similar to the viruses in the order Picornavirales and family Potyviridae in that they possess a genome-linked protein, VPg, at the 5′-terminus and a poly(A) tract at the 3′-terminus of the genome. The viral RNA-dependent RNA polymerase and the protease of caliciviruses share sequence homology with the picornaviruses.
Derivation of names
Calici: from Latin calix, “cup” or “goblet”, from cup-shaped depressions on the virion surface observed by electron microscopy.
Vesi: from the type species name vesicular exanthema of swine virus.
Lago: from Lagomorpha, the mammalian host order for the prototype strain rabbit haemorrhagic disease virus.
Noro: modified from the type species name Norwalk virus.
Sapo: modified from the type species name Sapporo virus.
Nebo: derived by using the first two letters from Newbury and Nebraska, the first two isolates to be studied together with bo for bovine the only host species from which these viruses have been described.
Further reading
Farkas et al., 2004 T. Farkas, W.M. Zhong, Y. Jing, P.W. Huang, S.M. Espinosa, N. Martinez, A.L. Morrow, G.M. Ruiz-Palacios, L.K. Pickering, X. Jiang, Genetic diversity among sapoviruses. Arch Virol. 149 (2004) 1309–1323.
Farkas et al., 2008 T. Farkas, K. Sestak, C. Wei, X. Jiang, Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J. Virol. 82 (2008) 5408–5416.
Glass et al., 2009 R.I. Glass, U.D. Parashar, M.K. Estes, Norovirus gastroenteritis. N. Engl. J. Med. 361 (2009) 1776–1785.
Green, 2007 K.Y. Green, D.M. Knipe, P.M. Howley, Caliciviridae: The NorovirusesFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott Williams & Wilkins, Wolters Kluwer Business, Philadelphia2007949–980.
Hansman et al., 2007 G.S. Hansman, T. Oka, K. Katayama, N. Takeda, Human sapoviruses: genetic diversity, recombination, and classification. Rev. Med. Virol. 17 (2007) 133–141.
L'homme et al., 2009 Y. L'homme, R. Sansregret, E. Plante-Fortier, A.M. Lamontagne, M. Ouardani, G. Lacroix, C. Simard, Genomic characterization of swine caliciviruses representing a new genus of Caliciviridae. Virus Genes. 39 (2009) 66–75.
Neill et al., 1995 J.D. Neill, R.F. Meyer, B.S. Seal, Genetic relatedness of the Caliciviruses: San Miguel sea lion and Vesicular exanthema of swine viruses constitute a single genotype within the Caliciviridae. J. Virol. 69 (1995) 4484–4488.
Oliver et al., 2006 S.L. Oliver, E. Asobayire, A.M. Dastjerdi, J.C. Bridger, Genomic characterization of the unclassified bovine enteric virus Newbury agent-1 (Newbury1) endorses a new genus in the family. Caliciviridae. Virology. 350 (2006) 240–250.
Reid et al., 2007 S.M. Reid, D.P. King, A.E. Shaw, N.J. Knowles, G.H. Hutchings, E.J. Cooper, A.W. Smith, N.P. Ferris, Development of a real-time reverse transcription polymerase chain reaction assay for detection of marine caliciviruses (genus Vesivirus). J. Virol. Methods. 140 (2007) 166–173.
Smiley et al., 2002 J.R. Smiley, K.O. Chang, J. Hayes, J. Vinje, L.J. Saif, Characterization of an enteropathogenic bovine calicivirus representing a potentially new calicivirus genus. J. Virol. 76 (2002) 10089–10098.
Wirblich et al., 1994 C. Wirblich, G. Meyers, V.F. Ohlinger, L. Capucci, U. Eskens, B. Haas, H.-J. Thiel, European brown hare syndrome virus: relationship to rabbit hemorrhagic disease virus and other caliciviruses. J. Virol. 68 (1994) 5164–5173.
Zheng et al., 2006 D.-P. Zheng, T. Ando, R.L. Fankhauser, R.S. Beard, R.I. Glass, S.S. Monroe, Norovirus classification and proposed strain nomenclature. Virology. 346 (2006) 312–323.
Contributed by
Clarke, I.N., Estes, M.K., Green, K.Y., Hansman, G.S., Knowles, N.J., Koopmans, M.K., Matson, D.O., Meyers, G., Neill, J.D., Radford, A., Smith, A.W., Studdert, M.J., Thiel, H.-J. and Vinjé, J.
Figures
Figure 1 The structure of the calicivirus capsid exemplified by cryo-image reconstruction of recombinant Norwalk virus (NV)-like particles (rNV VLPs) (Left). X-ray structure of the Norwalk virus capsid (Right) with the Shell, Protruding 1, and Protruding 2 domains colored in blue, red and yellow, respectively.
(Courtesy of B.V. Prasad.)

Figure 2 Reading frame usage and gene order in the Caliciviridae. Caliciviruses have a positive-strand RNA genome (7.48.3 kb), depicted by the horizontal black lines, which carry a protein (VPg) covalently linked to the 5-terminus. The genome also contains a characteristic, short repeated sequence () at the 5-terminus and at the start of the VP1 gene. The genome (of each genus as indicated) is organized into at least two or three major ORFs as indicated. Structural genes are shown as pink (VP1) and blue (VP2) and the VPg gene is shown as green. In all genera, the structural proteins VP1 and VP2 are produced during replication from an abundant subgenomic RNA transcript that is co-terminal with the 3-terminus of the genome. The ORF1 cleavage events mediated by the viral cysteine protease vary among the genera, but the gene order of the nonstructural proteins is conserved. Conserved cleavage sites are indicated with a solid vertical line; cleavage sites that vary in usage among genera are indicated with a broken vertical line.
(Re-designed by Rachel Skilton.)
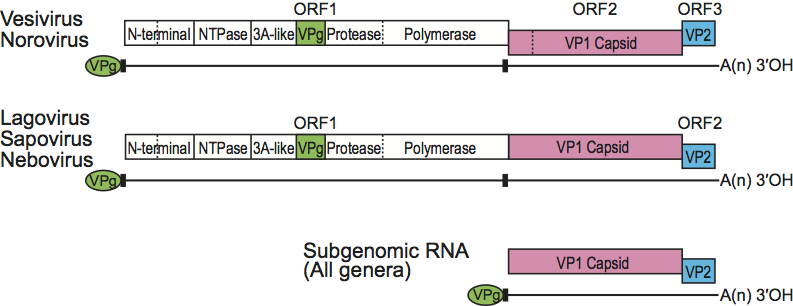
Figure 3 Phylogenetic relationships in the family Caliciviridae. The noroviruses are highlighted in red, lagoviruses in light blue, neboviruses in yellow, vesiviruses in green and sapoviruses in blue. Two prototype viruses were included to represent each genogroup, For some genera, additional strains were included to demonstrate circulation in different species (e.g., Po/Sw917 in norovirus GII). Genera are defined by amino acid p-distance of up to 0.7, genera cut-offs were expanded to include all representative genogroups within a genus. The amino acids of full length VP1 capsid sequences were aligned in SeaView 4.2 using MUSCLE with a gap cost of 5 and neighbor-joining clustering method for the 1st and 2nd iterations. Bayesian phylogenetic analysis was run on BEAST without a molecular clock for 106 iterations using the WAG amino acid substitution and Yule speciation tree model. Parameters and length of the run was verified by using Tracer, part of the BEAST suite of programs. All genera are supported by posterior distribution value of 1.0 (100% of sampled trees). The scale bar represents amino acid substitutions per site.
(Courtesy of Everardo Vega.)

