Family: Bromoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are either spherical or quasi-spherical (Figure 1), having T=3 icosahedral symmetry, and a diameter of 26–35 nm (genera Anulavirus, Bromovirus, Cucumovirus and Ilarvirus) or bacilliform (genera Alfamovirus, Ilarvirus and Oleavirus) having (within a species) constant diameters of 18–26 nm and lengths from 30 to 85 nm. Genomic RNAs are packaged in separate virions that may also contain sgRNAs, defective RNAs or satellite RNAs.
Physicochemical and physical properties
The Mr of the virions varies from 3.5–6.9×106, depending on the nucleic acid content. Virion Mr is constant among members of the genera Bromovirus, Cucumovirus and some Ilarvirus members, but varies among the remaining members of the family. The buoyant density of formaldehyde-fixed virions ranges from 1.35 to 1.37 g cm−3 in CsCl. The S20,w varies from 63S to 99S. Virion integrity is dependent on RNA–protein interactions and virion RNA is susceptible to RNAse degradation in situ at neutral pH. Heat inactivation occurs at 60 °C in some genera, others have not been tested. In some cases, virions are unstable in the presence of divalent cations. Virions are generally stable in the presence of chloroform, but not in the presence of phenol. Virions are unstable in the presence of strong anionic detergents such as SDS, but can tolerate low concentrations of mild detergents such as Triton X-100.
Nucleic acid
Total genome length is approximately 8 kb. Genomes consist of three linear, positive sense ssRNAs with 5′-terminal cap structures. The 3′ termini are not polyadenylated, but generally are highly conserved within a species or isolate, and form strong secondary structures. They are either tRNA-like and can be aminoacylated (genera Bromovirus and Cucumovirus) or form other structures that are not aminoacylated (genera Alfamovirus, Anulavirus, Ilarvirus and Oleavirus) (Table 1).
Table 1 Genomic RNA sizes in nucleotide number for the type members of each genus
| Genus | Species | Strain | RNA-1 | RNA-2 | RNA-3 | 3′ term. | DIs/sat RNAs |
| Alfamovirus | AMV | 425 | 3,644 | 2,593 | 2,037 | Complex* | −/− |
| Anulavirus | PZSV | Tomato | 3,383 | 2,435 | 2,659 | Complex | −/− |
| Bromovirus | BMV | Russian | 3,234 | 2,865 | 2,117 | tRNA† | +/− |
| Cucumovirus | CMV | Fny | 3,357 | 3,050 | 2,216 | tRNA | +/+ |
| Ilarvirus | TSV | N.A. | 3,491 | 2,926 | 2,205 | Complex | −/− |
| Oleavirus | OLV-2 | N.A. | 3,126 | 2,734 | 2,438 | Complex | ?/? |
N.A.: not applicable, only one strain reported.
* Complex secondary structure.
† Aminoacylatable, with pseudoknot folding.
Proteins
A single 20–24 kDa CP is expressed from a sgRNA. The CP is generally required for systemic movement and may be required for cell-to-cell spread in some cases.
Lipids
There are no lipids associated with the virions.
Carbohydrates
There are no carbohydrates associated with the virions.
Genome organization and replication
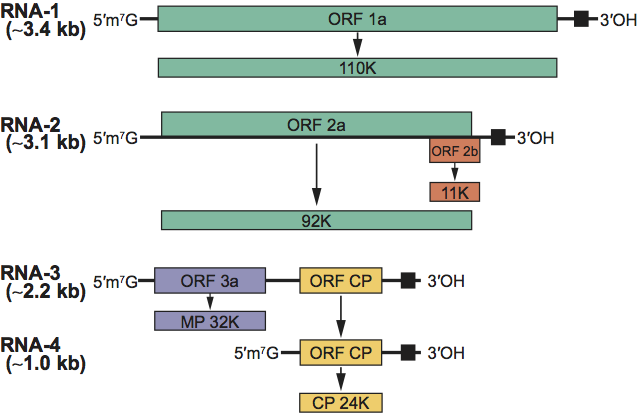
RNA-1, -2, and -3 can act as mRNAs. The genomic RNA-1 and -2 each encodes a single large ORF. These proteins (1a and 2a) act with host factors as the viral replicase. In the genus Cucumovirus and in some members of the genus Ilarvirus (subgroups 1 and 2 only) a smaller 2b protein is expressed from an additional sgRNA that may or may not be encapsidated. These 2b proteins are involved in cell-to-cell movement and post-transcriptional gene silencing. The 3a protein is a movement protein and ORF3b encodes the coat protein, expressed from a sgRNA (Figure 2; Table 2).
There is no clear evidence of proteolytic or other post-translational processing. Virus replication occurs on cytoplasmic membranes via full length minus (−) strand synthesis and subsequent plus (+) strand synthesis. The sgRNAs are synthesized from the (−) template, and may or may not be found in the virions. The CP accumulates to high levels in infected cells, whereas the nonstructural proteins accumulate to much lower levels. Virions accumulate in the cytoplasm. The life cycle of the virus takes place predominantly in the cytoplasm.
Table 2 Virus proteins, sizes and functional activities
| Protein | Size (kDa) | mRNA | Function* |
| 1a | 102.7–125.8 | RNA-1 | Mtr, helicase |
| 2a | 78.9–96.7 | RNA-2 | RdRp |
| 3a | 30.5–36.5 | RNA-3 | Cell to cell movement |
| CP | 19.8–26.2 | sgRNA-4† | Encapsidation, movement |
A 2b protein that is involved in cell-to-cell movement and post-transcriptional gene silencing is coded for by members of the genus Cucumovirus and some members of the genus Ilarvirus but is not found in other members of the genus Ilarvirus or in members of other genera within the family.
* Functions of 1a and 2a are putative in most cases, by analogy to related viruses.
† The sgRNA for the CP derived from RNA-3 is encapsidated in all but the genus Oleavirus.
Antigenic properties
Native virions are generally poor immunogens and require stabilization with formaldehyde prior to use as antigens. There are few or no serological relationships between the genera, and weak relationships between species of the same genus.
Biological properties
The natural host range of the viruses ranges from very narrow (genus Bromovirus) to extremely broad (genus Cucumovirus). They are predominantly transmitted by insects, in either a non-persistent manner or mechanically. Vectors have not been identified for some of the members of the family. They are distributed worldwide, and several are responsible for major disease epidemics in crop plants.
Genus Alfamovirus
Type species Alfalfa mosaic virus
Distinguishing features
Alfamoviruses are transmitted in a non-persistent manner by at least 14 aphid species. AMV shares many properties at the molecular level with members of the genus Ilarvirus. The CP is required to activate the genome, a feature shared with the ilarviruses. The CPs of AMV and ilarviruses can activate their respective genomes interchangeably.
Virion properties
Morphology
Virions are generally bacilliform, having a constant diameter of 18 nm, and varying from 30 to 57 nm in length, depending on the nucleic acid species encapsidated. The Mr values are from 3.5–6.9×106. Virions can be separated into components on sucrose density gradients.
Genome organization and replication
Replication is activated by CP binding to the complex 3′-end structure. CP from members of the genus Ilarvirus can also activate replication.
Biological properties
The host range is very broad. The virus is transmitted by aphids in a non-persistent manner.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Alfamovirus
| Alfalfa mosaic virus |
|
|
| Alfalfa mosaic virus - 425 | RNA1:[L00163=NC_001495] | (AMV-425) |
|
| RNA2:[X01572=NC_002024] |
|
|
| RNA3:[K02703=NC_002025] |
|
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Alfamovirus but have not been approved as species
| Cassava Ivorian bacilliform virus | (CIBV) |
Genus Anulavirus
Type species Pelargonium zonate spot virus
Distinguishing features
The RNA 3 is slightly larger than the RNA 2, unlike other members of this family. The 2a protein is the smallest reported for viruses in the family (78.9 kDa).
Virion properties
Morphology
Virus particles are non-enveloped and quasi-spherical, with a diameter ranging from 25 to 35 nm, and have a poorly resolved surface structure.
Physicochemical and physical properties
In sucrose density gradients, virions sediment as three components. In analytical centrifugation virions have single buoyant densities at equilibrium of 1.35 g cm−3 in CsCl or 1.29 g cm−3 in Cs2SO4.
Genome organization and replication
Coat protein is not required for activation of the genome.
Antigenic properties
Virions are weak immunogens and must be fixed before injection into rabbits to raise antibodies. Polyclonal antisera yield a single precipitin line in immuno-diffusion tests.
Biological properties
The single member of the genus infects tomato and artichoke and is common in weeds (e.g. Diplotaxis erucoides where it is also seed-borne). The virus is present in/on the pollen that is carried on the bodies of thrips feeding on susceptible hosts. Infected cells have severe cytopathological alterations.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Anulavirus
| Pelargonium zonate spot virus |
|
|
| Pelargonium zonate spot virus - tomato | RNA1:[AJ272327=NC_003649] | (PZSV-tomato) |
|
| RNA2:[AJ272328=NC_003650] |
|
|
| RNA3:[AJ272329=NC_003651] |
|
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Anulavirus but have not been approved as species
None reported.
Genus Bromovirus
Type species Brome mosaic virus
Distinguishing features
Beetle vectors are recorded for most bromoviruses but the efficiency of such transmission is low. A long-standing report of transmission by nematodes is regarded with suspicion.
Virion properties
Morphology
Virions are polyhedral, and all the same size, with a diameter of 27 nm.
Physicochemical and physical properties
Virions prepared below pH 6.0 have S20,w of 88S, are stable to high salt and low detergent concentrations, and are nuclease- and protease-resistant. At pH 7.0 and above, virions swell to a diameter of 31 nm, S20,w decreases to 78S, salt and detergent stability decreases dramatically, and protein and RNA are susceptible to hydrolytic enzymes. This swelling is accompanied by conformational changes of the capsid that are detectable by physical and serological means.
Nucleic acid
RNA 3′ termini are tRNA-like, are very similar in all viruses sequenced so far, and can be aminoacylated with tyrosine.
Antigenic properties
All members are serologically related, with large antigenic differences between species.
Biological properties
The natural host range is narrow, and is limited to a few plant hosts for each species. All species are thought to be beetle-transmitted, although BMV is inefficiently transmitted by aphids in a non-persistent manner.
Species demarcation criteria in the genus
Criteria used for demarcation of species within the genus are:
- Host range
- Serological relationships
- Compatible replicase proteins (i.e. 1a and 2a proteins)
- Nucleotide sequence identity between species ranges from 50 to 80% depending on the gene used for comparison.
List of species in the genus Bromovirus
| Broad bean mottle virus |
|
|
| Broad bean mottle virus - Bawden | RNA1:[M65138=NC_004008] | (BBMV-Bawden) |
|
| RNA2:[M64713=NC_004007] |
|
|
| RNA3:[M60291=NC_004006] |
|
| Brome mosaic virus |
|
|
| Brome mosaic virus - Russian | RNA1:[X02380=NC_002026] | (BMV-RU) |
|
| RNA2:[X01678=NC_002027] |
|
|
| RNA3:[J02042=NC_002028] |
|
| Cassia yellow blotch virus |
|
|
| Cassia yellow blotch virus - KU1 | RNA1:[AB194806=NC_006999] | (CYBV-KU1) |
|
| RNA2:[AB194807=NC_007000] |
|
|
| RNA3:[AB194808=NC_007001] |
|
| Cowpea chlorotic mottle virus |
|
|
| Cowpea chlorotic mottle virus – type | RNA1:[M65139=NC_003543] | (CCMV-Type) |
|
| RNA2:[M28817=NC_003541] |
|
|
| RNA3:[M28818=NC_003542] |
|
| Melandrium yellow fleck virus |
|
|
| Melandrium yellow fleck virus - KU1 | RNA1:[AB444583=NC_013266] | (MYFV –KU1) |
|
| RNA2:[AB444584=NC_013267] |
|
|
| RNA3:[AB444585=NC_013268] |
|
| Spring beauty latent virus |
|
|
| Spring beauty latent virus - KU1 | RNA1:[AB080598=NC_004120] | (SBLV-KU1) |
|
| RNA2:[AB080599=NC_004121] |
|
|
| RNA3:[AB080600=NC_004122] |
|
Species names are in italic script; isolate and strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Bromovirus but have not been approved as species
None reported.
Genus Cucumovirus
Type species Cucumber mosaic virus
Distinguishing features
Cucumoviruses are transmitted in a non-persistent manner by over 80 species of aphids in more than 30 genera. The RNA 2 is bistronic producing a 2b protein associated with the suppression of RNA interference.
Virion properties
Morphology
Virions are icosahedral, of uniform size and sedimentation properties. In electron micrographs they appear to have electron dense centers.
Physicochemical and physical properties
Purified virions are labile, and are especially susceptible to anionic detergents and high ionic strength buffers that disrupt the RNA-protein interactions required for particle integrity. Most strains are unstable in the presence of Mg2+, but at least one strain of cucumber mosaic virus (CMV) requires Mg2+ for stability.
Nucleic acid
The 3′ termini of all RNAs within a species are highly similar and can form tRNA-like structures that are aminoacylatable with tyrosine. Within a species, the 5′-NTR of RNA-1 and RNA-2 are also very similar. At least one strain of CMV can form defective RNAs that arise by deletions in the 3a ORF of RNA-3. Subgroup II strains of CMV encapsidate the sgRNA for the 2b ORF, called RNA-4a, and an additional small RNA of about 300 nt, called RNA-5, that is co-terminal with the 3′ ends of RNAs-3 and 4. In addition, CMV and peanut stunt virus (PSV) may harbor satellite RNAs of about 330 to 400 nt. The satellite RNAs are more common under experimental conditions than in field conditions, and may dramatically alter the symptoms of infection by the helper virus in certain hosts like tomato.
Genome organization and replication
An additional ORF, the 2b ORF, is found in all cucumoviruses and has been shown to be active in CMV and tomato aspermy virus (TAV).
Antigenic properties
CMV has been divided into two subgroups, based on serology. PSV also has more than one serological group. Sequence analysis has upheld the divisions, although the CMV subgroup I can be further divided into two groups by phylogenetic analyses.
Biological properties
CMV has an extremely broad host range, infecting 85 distinct plant families, and up to 1000 species experimentally. The other cucumovirus species have narrower host ranges; PSV is largely limited to legumes and solanaceous hosts, and TAV predominantly infects composites and solanaceous plants. All species are transmitted by aphids in a non-persistent manner.
Species demarcation criteria in the genus
Criteria used for demarcation of species within the genus are:
- Serological relatedness
- Compatibility of replicase proteins (1a and 2a proteins), but these distinctions may break down in the case of naturally occurring reassortants
- Sequence similarity.
Serology and nucleotide sequence similarity is used to distinguish subgroups within a species. Subgroups generally have at least 65% sequence identity.
List of species in the genus Cucumovirus
| Cucumber mosaic virus |
|
|
| Cucumber mosaic virus subgroup I strain Fny | RNA1:[D00356=NC_002034] | (CMV-Fny) |
|
| RNA2:[D00355=NC_002035] |
|
|
| RNA3:[D10538=NC_001440] |
|
| Cucumber mosaic virus subgroup II strain Q | RNA1:[X02733] | (CMV-Q) |
|
| RNA2:[D00985] |
|
|
| RNA3:[M21464] |
|
| Peanut stunt virus |
|
|
| (Robinia mosaic virus) |
|
|
| Peanut stunt virus - ER | RNA1:[U15728=NC_002038] | (PSV-ER) |
|
| RNA2:[U15729=NC_002039] |
|
|
| RNA3:[U15730=NC_002040] |
|
| Tomato aspermy virus |
|
|
| (Chrysanthemum aspermy virus) |
|
|
| Tomato aspermy virus - V | RNA1:[D10044=NC_003837] | (TAV-V) |
|
| RNA2:[D10663=NC_003838] |
|
|
| RNA3:[AJ277268=NC_003836] |
|
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Cucumovirus but have not been approved as species
| Gay feather mild mottle virus | RNA1:[FM881899=NC_012134] | (GMMV) |
|
| RNA2:[FM881900=NC_012135] |
|
|
| RNA3:[FM881901=NC_012136] |
|
Genus Ilarvirus
Type species Tobacco streak virus
Distinguishing features
Ilarviruses have been demonstrated to be transmitted mechanically by thrips feeding on pollen grains containing the virus. The RNA-2 of members of subgroups 1 and 2 is bicistronic producing a 2b protein which is inferred, because of functional homology, to serve the same function as the 2b protein of cucumoviruses.
Virion properties
Morphology
Virions are quasi-isometric or occasionally bacilliform, and are about 30 nm in diameter.
Nucleic acid
There is a short region of sequence similarity at the 3′ ends of the RNAs.
Genome organization and replication
The CP is required for activation of replication, but may be substituted with CP from alfalfa mosaic virus (genus Alfamovirus).
Antigenic properties
Virions are “unpromising subjects for the raising of good antisera”. Subgroups within the genus were based on the presence of serological relationships among some, but not all, members of each subgroup. Sequence data have supported many of the serological relationships but also show potential relationships among viruses not previously demonstrated as related. Some serological relationships have been reported between viruses in different subgroups and might be associated with conserved aa and secondary structures.
Biological properties
The viruses mainly infect woody plants. Viruses are present in/on the pollen and may be transmitted when wind-blown pollen and populations of vector thrips are coincident on a susceptible host.
Species demarcation criteria in the genus
Criteria used for demarcation of species within the genus are:
- Serology
- Host range
- Sequence similarity (Specific levels of sequence similarity have not been defined).
Ilarviruses were initially grouped on the basis of serological relationships. Sequence data have confirmed some of these groupings but have also shown that some species were grouped inappropriately. Currently accepted groupings of the viruses are indicated in the species list.
List of species in the genus Ilarvirus
| Subgroup 1 |
|
|
| Parietaria mottle virus |
|
|
| Parietaria mottle virus – Caciagli | RNA1:[AY496068=NC_005848] | (PMoV - Caciagli) |
|
| RNA2:[AY496069=NC_005849] |
|
|
| RNA3:[U35145=NC_005854] |
|
| Tobacco streak virus |
|
|
| Tobacco streak virus – WC | RNA1:[U80934=NC_003844] | (TSV-WC) |
|
| RNA2:[U75538=NC_003845] |
|
|
| RNA3:[X00435=NC_002028] |
|
| Subgroup 2 |
|
|
| Asparagus virus 2 |
|
|
| Asparagus virus 2 - Mink | RNA1:[EU919666=NC_011808] | (AV-2 - Mink) |
|
| RNA2:[EU919667=NC_011809] |
|
|
| RNA3:[X86352=NC_011807] |
|
| Citrus leaf rugose virus |
|
|
| Citrus leaf rugose virus – Garnsey | RNA1:[U23715=NC_003548] | (CiLRV - Garnsey) |
|
| RNA2:[U17726=NC_003547] |
|
|
| RNA3:[U17390=NC_003546] |
|
| Citrus variegation virus |
|
|
| Citrus variegation virus - Garnsey | RNA1:[EF584664=NC_009537] | (CVV - Garnsey) |
|
| RNA2:[EF584865=NC_009538] |
|
|
| RNA3:[U17389=NC_09536] |
|
| Elm mottle virus |
|
|
| (Hydrangea mosaic virus) |
|
|
| Elm mottle virus - Jones |
|
|
|
| RNA1:[U57047=NC_003569] | (EMoV - Jones) |
|
| RNA2:[U34050=NC_003568] |
|
|
| RNA3:[U85399=NC_003570] |
|
| Lilac ring mottle virus |
|
|
| Lilac ring mottle virus - Netherlands | RNA1:[EU919668] | (LiRMoV -Netherlands) |
|
| RNA2:[EU919669] |
|
|
| RNA3:[U17391] |
|
| Spinach latent virus |
|
|
| Spinach latent virus - Bos | RNA1:[U93192=NC_003808] | (SpLV - Bos) |
|
| RNA2:[U93193=NC_003809] |
|
|
| RNA3:[U93194=NC_003810] |
|
| Tulare apple mosaic virus |
|
|
| Tulare apple mosaic virus - Garnsey | RNA1:[AF226160=NC_003833] | (TaMV - Garnsey) |
|
| RNA2:[AF226161=NC_003834] |
|
|
| RNA3:[AF226162=NC_003835] |
|
| Subgroup 3 |
|
|
| Apple mosaic virus |
|
|
| Apple mosaic virus | RNA1:[AF174584=NC_003464] | (ApMV) |
|
| RNA2:[AF174585=NC_003465] |
|
|
| RNA3:[U15608=NC_003480] |
|
| Blueberry shock virus |
|
|
| Blueberry shock virus |
| (BlShV) |
| Prunus necrotic ringspot virus |
|
|
| Prunus necrotic ringspot virus | RNA1:[AF278534=NC_004362] | (PNRSV) |
|
| RNA2:[AF278535=NC_004363] |
|
|
| RNA3:[U57046=NC_004364] |
|
| Subgroup 4 |
|
|
| Fragaria chiloensis latent virus |
|
|
| Fragaria chiloensis latent virus – Tzanetakis | RNA1:[AY682102=NC_006566] | (FCiLV-Tzanetakis) |
|
| RNA2:[AY707771=NC_006567] |
|
|
| RNA3:[AY707772=NC_006568] |
|
| Prune dwarf virus |
|
|
| Prune dwarf virus | RNA1:[U57648=NC_008039] | (PDV) |
|
| RNA2:[AF277662=NC_008037] |
|
|
| RNA3:[L28145=NC_008038] |
|
| No relationships to other existing groups |
|
|
| American plum line pattern virus |
|
|
| American plum line pattern virus - Parrish | RNA1:[AF235033=NC_003451] | (APLPV -Parrish) |
|
| RNA2:[AF235165=NC_003452] |
|
|
| RNA3:[AF235166=NC_003453] |
|
| Humulus japonicus latent virus |
|
|
| Humulus japonicus latent virus – Adams | RNA1:[AY500236=NC_006064] | (HJLV - Adams) |
|
| RNA2:[AY500237=NC_006065] |
|
|
| RNA3:[AY500238=NC_006066] |
|
Species names are in italic script; names of strains and isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Ilarvirus but have not been approved as species
The potential subgroups to which these viruses will be assigned, once complete information and necessary approval as new species has been completed, are indicated.
| Subgroup 1 |
|
|
| Bacopa chlorosis virus | RNA1:[FJ607140] | (BaCV) |
|
| RNA2:[FJ607141] |
|
|
| RNA3:[FJ607142] |
|
| Blackberry chlorotic ringspot virus | RNA1:[DQ091193=NC_011553] | (BCRV) |
|
| RNA2:[DQ091194=NC_011554] |
|
|
| RNA3:[DQ091195=NC_011555] |
|
| Strawberry necrotic shock virus | RNA1:[DQ318818=NC_008708] | (SNSV) |
|
| RNA2:[AY743591=NC_008707] |
|
|
| RNA3: [AY363228=NC_008706] |
|
| Tomato necrotic spot virus | RNA3:[FJ236810] | (ToNSV) |
| Subgroup 3 |
|
|
| Lilac leaf chlorosis virus | RNA2:[FN669168] | (LLCV) |
|
| RNA3:[FN669169] |
|
| Subgroup 4 |
|
|
| Viola white distortion virus | RNA3:[GU168941] | (VWDV) |
Genus Oleavirus
Type species Olive latent virus 2
Distinguishing features
A fourth RNA with no apparent messenger activity, and which is slightly smaller than the RNA-3, is encapsidated by the virus.
Virion properties
Morphology
Virions have different shape and size, ranging from quasi-spherical with a diameter of 26 nm, to bacilliform with lengths of 37, 43, 38, and 55 nm, and diameters of 18 nm. Particles up to 85 nm in length occasionally are present.
Physicochemical and physical properties
In sucrose gradients, virions sediment as five or six components.
Nucleic acid
Virion RNA differs in size from that of other members of the family, encapsidating the three genomic RNAs and a sgRNA of about 2 kb, that is apparently not an mRNA. The sgRNA for the CP ORF is not encapsidated. Three additional RNAs of 200 to 550 nt are also present in virions. The 5′ termini of the genomic RNAs are capped, but not the 5′ terminus of the encapsidated sgRNA. The 3′ termini of the RNAs are similar to those of the genera Alfamovirus and Ilarvirus, but do not interact with CP to activate replication.
Antigenic properties
Virions are efficient immunogens.
Biological properties
The only known host is olive (Olea europaea), which is infected asymptomatically. The virus is transmitted by inoculations, but no insect vector is known.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Oleavirus
| Olive latent virus 2 |
|
|
| Olive latent virus 2 – Italy | RNA1:[X94346=NC_003673] | (OLV-2-Italy) |
|
| RNA2:[X94347=NC_003674] |
|
|
| RNA3:[X76993=NC_003671] |
|
Species names are in italic script; isolate names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Oleavirus but have not been approved as species
None reported.
Phylogenetic relationships within the family
Relationships within this family have been examined using the sequence of full-length genomic molecules, genes and gene products. The three genera with multiple members (Bromovirus, Cucumovirus and Ilarvirus) form clades that are distinct. The genera Anulavirus and Oleavirus are unique and distinct from other genera within the family. A longstanding debate as to whether AMV (genus Alfamovirus) should be regarded as a member of the genus Ilarvirus has yet to be resolved. Ilarviruses share many features at the molecular level with AMV. The CP of AMV will activate replication of ilarviruses and vice versa. The major difference between AMV and ilarviruses is in the mode of transmission. AMV is transmitted non-persistently by aphids and ilarviruses are transmitted through pollen and feeding of thrips. Studies defining recombination events in the genomes of members of the Bromoviridae suggest that AMV should be grouped with the ilarviruses.
Similarity with other taxa
The viruses are members of the “alpha-like” supergroup, sharing sequence similarity in the 1a protein domains for Mtr and Hel activities, and in the 2a protein polymerase domain with members of the plant virus family Virgaviridae and order Tymovirales, and animal viruses in the family Togaviridae. The 3a proteins of bromoviruses and the 35 kDa protein of the members of the genus Dianthovirus (RCNMV) form a distinct “family” of movement-associated proteins. Raspberry bushy dwarf virus (RBDV) (genus Idaeovirus) is similar to bromoviruses in genome organization and in the sequence of certain genes. Like the members of the genus Ilarvirus, RBDV is transmitted in association with pollen.
Derivation of names
Alfamo: from alfalfa mosaic virus.
Anula: from Latin “anular” for the concentric symptom associated with infection by this virus.
Bromo: from Brome mosaic, also, from Bromus (host of Brome mosaic virus).
Cucumo: from cucumber mosaic virus.
Ilar: from isometric labile ringspot.
Olea: from the genus name of the host, olive (Olea).
Further reading
See articles on this family and genera within the following sources:
The Springer Index of Viruses, 2011 The Springer Index of Viruses (Tidona, C.A. and Darai, G., Eds.), Springer Verlag, Berlin, 2001 and 2011.
The Encyclopedia of Virology, 2008 The Encyclopedia of Virology (Mahey, B.W.J. and van Regenmortel, M.H.V., Eds.), Elsevier, Amsterdam, 2008.
The Encyclopedia of Life Sciences, 2010 The Encyclopedia of Life Sciences, Wiley Interscience, New York, 2010.
Codoñer and Elena, 2008 F.M. Codoñer, S.F. Elena, The promiscuous evolutionary history of the Bromoviridae. J Gen. Virol. 89 (2008) 1739–1747.
Gallitelli et al., 2005 D. Gallitelli, M.M. Finetti Sialer, G.P. Martelli, Anulavirus a proposed new genus of plant viruses in the family Bromoviridae. Arch. Virol. 150 (2005) 407–411.
Martelli and Grieco, 1997 G.P. Martelli, F. Grieco, Oleavirus, a new genus in the family Bromoviridae. Arch. Virol. 142 (1997) 1933–1936.
Palukaitis and García-Arenal, 2003 P. Palukaitis, F. García-Arenal, Cucumoviruses. Adv. Virus. Res. 62 (2003) 241–323.
Scott et al., 2003 S.W. Scott, M.T. Zimmerman, X. Ge, Viruses in subgroup 2 of the genus Ilarvirus share both serological relationships and characteristics at the molecular level. Arch. Virol. 148 (2003) 2063–2075.
Contributed by
Bujarski, J., Figlerowicz, M., Gallitelli, D., Roossinck, M.J. and Scott, S.W.
Figures
Figure 1 (Top left) Image reconstruction of a particle of cowpea chlorotic mottle virus (CCMV) (genus Bromovirus), showing pentamer and hexamer clustering in a T=3 quasi-icosahedron (from Lucas et al. (2001). J. Mol. Biol., 317, 95-108). (Top central) schematic representation of a T=3 particle; (top right) negative contrast electron micrograph of particles of cucumber mosaic virus (CMV) (genus Cucumovirus) (courtesy of G. Kasdorf). (Bottom left) Electron density representation of a Ta particle of alfalfa mosaic virus (AMV) (genus Alfamovirus), showing T=1 structure (from Kumar et al. (1997). J. Virol., 71, 7911-7916). (Bottom center) Schematic representation of a T=1 particle. (Bottom right) Negative contrast electron micrograph of particles of prune dwarf virus (PDV) (genus Ilarvirus) (courtesy of G. Kasdorf). The bars represent 100 nm.
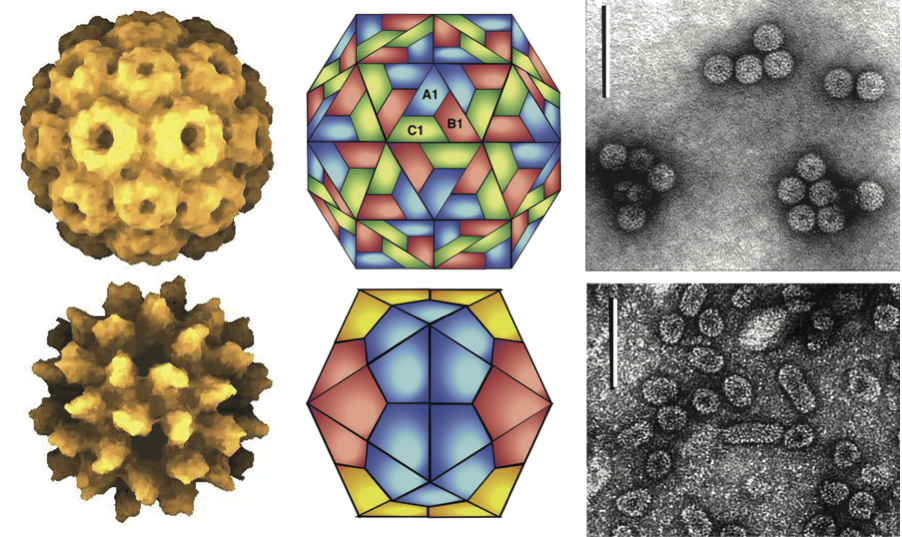
Figure 2 Schematic genome organization for members of the family Bromoviridae.