Genus: Benyvirus
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Type species: Beet necrotic yellow vein virus
Distinguishing features
Multipartite rod-shaped viruses containing positive stranded RNA genomes with 5′ m7G cap and 3′ polyA and post-translational cleavage of the viral replicase.
Virion properties
Morphology
Viral particles consist of four to five non-enveloped, helically constructed rod-shaped structures, with an axial canal (Figure 1). They have predominant lengths of about 390, 265, 100, 85 and 80 to 65 nm and a constant diameter of 20 nm. The right-handed helix with a pitch of 2.6 nm has an axial repeat of four turns, involving 49 CP subunits, each occupying four nucleotides. Coat proteins constitute about 95% of the particle weight.
Physicochemical and physical properties
The viral particles are sometimes unstable in sap with strong infectivity losses within one to five days at room temperature.
Nucleic acid
In naturally infected plants, viruses contain four to five linear positive sense ssRNAs of about 6.7, 4.6, 1.8, 1.4 and 1.3 kb, respectively. The viral RNAs are capped at the 5′ end and, unlike the RNAs of all other rod-shaped plant viruses, are 3′-polyadenylated. Viral RNAs harbour a conserved 3′ structure essential for RNA replication initiation. After mechanical transmission to laboratory test plants, beet necrotic yellow vein virus (BNYVV) RNAs 3, 4 and 5 may carry deletions or may be lost entirely. The complete sequence has been determined for all five RNAs of different isolates of BNYVV and for the four RNAs of an isolate of beet soil-borne mosaic virus (BSBMV). The possible additional members rice stripe necrosis virus (RSNV) and burdock mottle virus (BdMV) apparently only contain two genomic RNAs.
Proteins
The major CP species is 21–23 kDa in size. The minor CP amber readthrough protein is detected near one extremity of BNYVV particles and initiates virus assembly. The minor capsid protein contains a KTER motif in its C terminal part that is necessary for the transmission of the virions by Polymyxa betae.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
RNA-1 contains one large ORF coding for a replication-associated protein that is cleaved post-translationally. This proteolytic cleavage of the replicase distinguishes the benyviruses from all other viruses with rod-shaped particles, which have their replication-associated proteins encoded on two ORFs. In vitro translation of BNYVV RNA-1 may initiate at two sites: at the first AUG in the sequence at position 154 or at a downstream AUG at position 496. The resulting proteins of 237 and 220 kDa, respectively, both contain in their N-terminal part Mtr motifs, in their central part Hel and papain-like protease motifs (Prot) and in their C-terminal part RdRp motifs (Figure 2). RNA-2 of BNYVV contains six ORFs, i.e. the CP gene which is terminated by a suppressible amber stop codon (UAG), the CP readthrough protein gene, the TGB coding for TGB1, TGB2 and TGB3 (42, 13 and 15 kDa respectively) and a gene coding for P14 cysteine-rich protein, a viral posttranscriptional gene-silencing suppressor (14 kDa). The TGB ORFs and the P14 gene are expressed by means of subgenomic RNAs (Figure 2). The three triple gene block (TGB)-encoded proteins (TGB1, TGB2 and TGB3) but not the CP are necessary for cell-to-cell movement but long distance movement requires CP. BNYVV TGB1 labeled with green fluorescent protein (GFP) on its N-terminus is targeted by TGB2 and TGB3 to punctate bodies associated with plasmodesmata. The N-terminal part of BNYVV TGB1 has nucleic acid binding activity and its C-terminal part contains consensus sequence motifs characteristic of an ATP/GTP-dependent helicase. The three proteins are found along membrane-rich peripheral bodies, thought to be derived from endoplasmic reticulum (ER). The MP of tobacco mosaic virus can functionally complement TGB proteins. RNAs 1 and 2 are sufficient for replication of BNYVV in the local lesion host Chenopodium quinoa or in the systemic host Spinacia olacera. The typical rhizomania symptoms in beet are produced only in the presence of RNA-3; RNA-4 greatly increases the transmission rate by Polymyxa betae and RNA-5 may modulate the type of symptoms formed. RNAs-3 and -4 are always present in natural BNYVV infections. BNYVV is able to replicate and encapsidate BSBMV RNA-3.
Antigenic properties
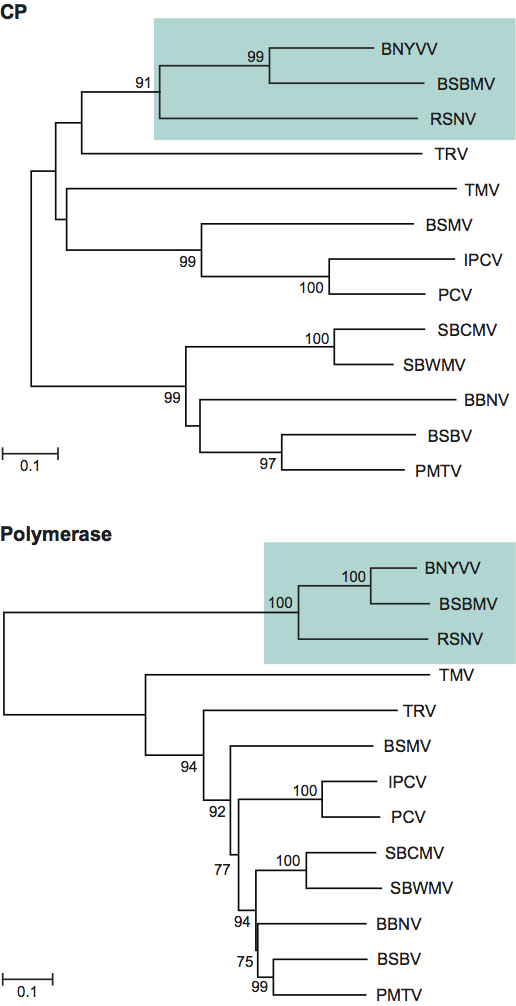
BNYVV and BSBMV are distinct serologically. Epitope mapping has revealed portions of the amino acid sequence of BNYVV CP that are either exposed along the entire particle length, e.g. the immunodominant C-terminus, or are accessible only on one extremity or after disruption of the particles (Figure 1).
Biological properties
BNYVV has been found at the cytoplasmic surface of mitochondria soon after infection. Later, virions of most BNYVV isolates are scattered throughout the cytoplasm of infected cells or occur in aggregates. More or less dense masses of particles arranged in parallel or angle-layer arrays may be formed. Depending on the isolate only one or both types of aggregates occur. Membranous accumulations of ER may also be found. The p25 protein and its tetrad sequence variability appear involved in the resistance breaking capabilities of the virus on sugar beet.
The natural host ranges of benyviruses are very narrow. Beta vulgaris is the natural host of BNYVV and BSBMV. Species of Chenopodium are infected experimentally, often only locally. Nicotiana benthamiana, Spinacia olacera and Beta macrocarpa are systemic hosts for BNYVV and BSBMV. In nature BNYVV and BSBMV are transmitted by Polymyxa betae and RNSV by P. graminis. The viruses are also mechanically transmissible.
BNYVV has spread to most sugar beet-growing areas worldwide. Variants (A-, B- or P-type) occur in different geographical areas and are distinguished by minor changes in the CP sequence, four amino acids in the p25 ORF (tetrad) and the presence of the RNA-5 species. BSBMV is widely distributed in the United States. RSNV occurs in Africa, and South and Central America. BdMV has been identified in a restricted area in Japan.
Species demarcation criteria in the genus
Benyviruses are distinguished by coat protein sequence less than 90% identical, distant serological relations and distinct host ranges.
List of species in the genus Benyvirus
| Beet necrotic yellow vein virus |
|
|
| Beet necrotic yellow vein virus - F2, France (B type) | [X05147; X04197; M36894; M36896] | (BNYVV-B-Eur) |
| Beet necrotic yellow vein virus - S, Japan (A type) | [D84410; D84411; D84412; D84413; D63936] | (BNYVV-A-Jap) |
| Beet necrotic yellow vein virus - F72, France (P type) | [HM126464; HM117903; DQ682454; DQ682453; AY823407] | (BNYVV-P-Eur) |
| Beet soil-borne mosaic virus |
|
|
| Beet soil-borne mosaic virus - EA | [AF280539; AF061869; AF280540; AF280541] | (BSBMV-EA) |
| Beet soil-borne mosaic virus - MRM06,* USA | [EU410955; FJ424610] | (BSBMV-MRM06) |
Species names are in italic script; names of strains and isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Complete RNA3 and 4 sequences only.
List of other related viruses which may be members of the genus Benyvirus but have not been approved as species
| Burdock mottle virus |
| (BdMV) |
| Rice stripe necrosis virus | [EU099844; EU099845] | (RSNV) |
Phylogenetic relationships within the genus
The CP amino acid sequences of BNYVV and BSBMV share less than 60% homology (Figure 3). The percentages of identity between various non-structural proteins of the two viruses range between 38% (cysteine-rich protein) and 84% (replication-associated protein). RNA species of BNYVV and BSBMV share common structural motifs at their extremities but coding sequences differ. BNYVV p26 protein resembles BSBMV p29 (Figure 2) in sequence and functions. The “coremin” sequence of 20 nucleotides involved in the systemic movement of the virus is present on: BNYVV RNA-3 core sequence and RNA-5, BSBMV RNA-3 and RNA-4 (Figure 2) as well as on cucumber mosaic virus RNAs. The CP of BdMV shares 38% sequence identity with that of BNYVV. TGB1 of BdMV shares c. 50% identity with the corresponding proteins of BNYVV and BSBMV. A 2,239 nucleotide fragment of RSNV RNA-1 shares about 50% sequence identity with the corresponding regions of BNYVV and BSBMV RNA-1.
Similarity with other taxa
Benyviruses are morphologically similar to other rod-shaped viruses that are classified in the family Virgaviridae (furo-, peclu-, pomo-, hordei-, tobra- and tobamoviruses). Conserved residues are found among the CP of these viruses, e.g. RF and FE in their central and C-terminal parts, respectively, which are presumably involved in the formation of salt bridges. Like pomo-, peclu- and hordeiviruses, but unlike furo-, tobamo- and tobraviruses, the benyviruses have their movement function encoded on a triple gene block. Sequence identities in the first and second triple gene block-encoded proteins reveal affinities not only to pomo- and hordei-, but also to potex- and carlaviruses. The Mtr, Hel and RdRp motifs in the replication-associated proteins show a higher degree of similarity to those of the viruses of the family Togaviridae (rubella virus) and of hepatitis virus E than to those of other rod-shaped plant viruses.
Derivation of name
Beny: from beet necrotic yellow vein virus.
Further reading
Acosta-Leal et al., 2010 R. Acosta-Leal, B.K. Bryan, J.T. Smith, C.M. Rush, Breakdown of host resistance by independent evolutionary lineages of Beet necrotic yellow vein virus involves a parallel c/u mutation in its p25 gene. Phytopathol. 100 (2010) 127–133.
Commandeur et al., 1994 U. Commandeur, R. Koenig, R. Manteuffel, L. Torrance, P. Lüddecke, R. Frank, Location, size and complexity of epitopes on the coat protein of beet necrotic yellow vein virus studied by means of synthetic overlapping peptides. Virology. 198 (1994) 282–287.
Erhardt et al., 2005 M. Erhardt, G. Vetter, D. Gilmer, S. Bouzoubaa, K. Richards, G. Jonard, H. Guilley, Subcellular localization of the Triple Gene Block movement proteins of Beet necrotic yellow vein virus by electron microscopy. Virology. 340 (2005) 155–166.
Haeberlé et al., 1994 A.M. Haeberlé, C. Stussi-Garaud, C. Schmitt, J.C. Garaud, K.E. Richards, H. Guilley, G. Jonard, Detection by immunogold labelling of P75 readthrough protein near an extremity of Beet necrotic yellow vein virus particles. Arch. Virol. 134 (1994) 195–203.
Hehn et al., 1997 A. Hehn, C. Fritsch, K.E. Richards, H. Guilley, G. Jonard, Evidence for in vitro and in vivo autocatalytic processing of the primary translation product of Beet necrotic yellow vein virus RNA 1 by a papain-like proteinase. Arch. Virol. 142 (1997) 1051–1058.
Lozano and Morales, 2009 I. Lozano, F. Morales, Molecular characterisation of Rice stripe necrosis virus as a new species of the genus Benyvirus. Eur. J. Plant Pathol. 124 (2009) 673–680.
Peltier et al., 2008 C. Peltier, K. Hleibieh, H. Thiel, E. Klein, C. Bragard, D. Gilmer, Molecular biology of the Beet necrotic yellow vein virus. Plant Viruses. 2 (2008) 14–24.
Rahim et al., 2007 M.D. Rahim, I.B. Andika, C. Han, H. Kondo, T. Tamada, RNA4-encoded p31 of beet necrotic yellow vein virus is involved in efficient vector transmission, symptom severity and silencing suppression in roots. J. Gen. Virol. 88 (2007) 1611–1619.
Ratti et al., 2009 C. Ratti, K. Hleibieh, L. Bianchi, A. Schirmer, C. Rubies Autonell, D. Gilmer, Beet soil-borne mosaic virus RNA-3 is replicated and encapsidated in the presence of BNYVV RNA-1 and -2 and allows long distance movement in Beta macrocarpa. Virology. 385 (2009) 392–399.
Valentin et al., 2005 C. Valentin, P. Dunoyer, G. Vetter, C. Schalk, A. Dietrich, S. Bouzoubaa, Molecular basis for mitochondrial localization of viral particles during beet necrotic yellow vein virus infection. J. Virol. 79 (2005) 9991–10002.
Contributed by
Gilmer, D. and Ratti, C.
Figures
Figure 1 (Left) Scheme showing the accessibility to antibodies of various parts of the coat protein amino acid (aa) sequence in particles of beet necrotic yellow vein virus (BNYVV). Encircled numbers designate different epitopes. (Center) Negative contrast electron micrograph of stained purified particles of BNYVV. (Right) From left (a) negative contrast electron micrograph of a BNYVV particle and (b, c, d) computer-filtered micrographs of BNYVV particles (courtesy of A.C. Steven, from Virology113, 428 (1981); with permission). The bar represents 100 nm.
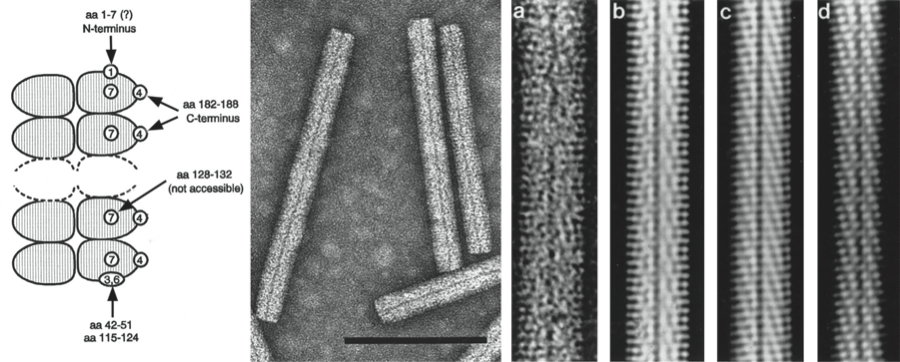
Figure 2 Genome organization and translation strategies of beet necrotic yellow vein virus (BNYVV) and beet soil-borne mosaic virus (BSBMV). The scheme indicates a self-cleavage of the replicase protein (red arrow and black triangle), a suppressible UAG stop codon (yellow star), m7Gppp (black circle) and (An) the 3 poly (A)-tails. Mtr, methyltransferase; Hel, helicase; Pro, protease; Pol, RNA polymerase; RT, readthrough; RTD, readthrough domain; sub, subgenomic; CRP, cysteine rich protein. N and p4.6 have never been detected. Nothing is known about BSBMV putative p11 and p13.
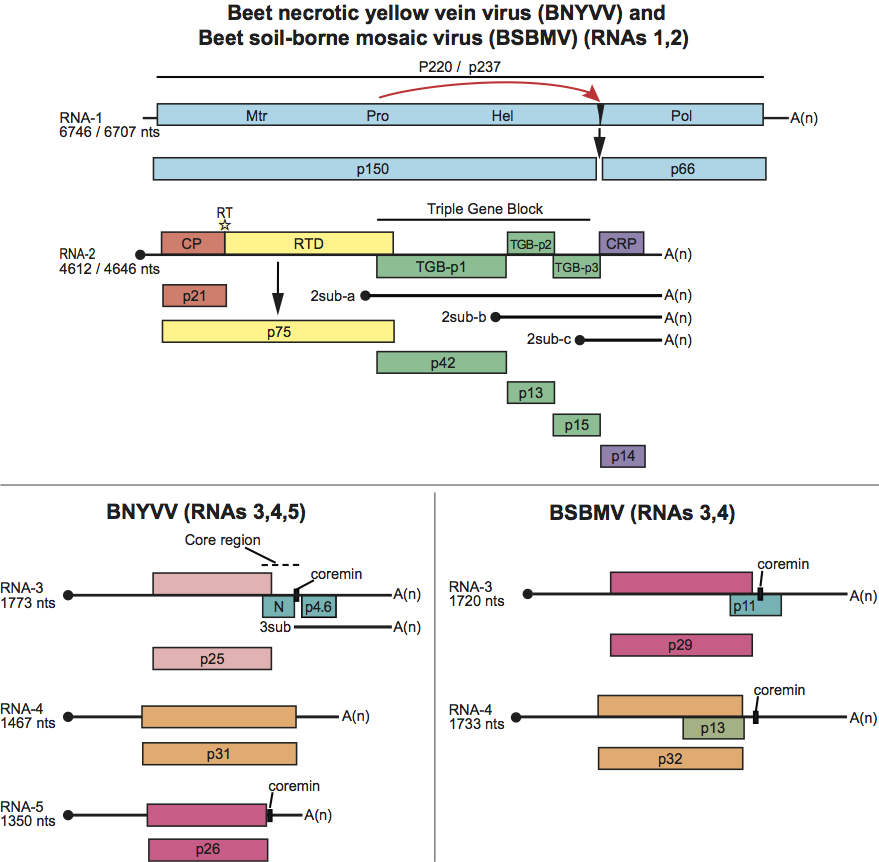
Figure 3 Phylogenetic (distance) tree based on the codon-aligned nucleotide sequences of the capsid (CP) and polymerase proteins of rod-shaped plant viruses. Numbers on branches indicate percentage of bootstrap support out of 10,000 bootstrap replications (when >60%). The scales indicate maximum composite likelihood distances. Trees produced in MEGA4. Viruses (or possible members) of the genus Benyvirus are highlighted. Other viruses are Genus Pomovirus: potato mop top virus (PMTV), beet soil-borne virus (BSBV), broad bean necrosis virus (BBNV); Furovirus: soil-borne cereal mosaic virus (SBCMV), soil-borne wheat mosaic virus (SBWMV); Tobamovirus: tobacco mosaic virus (TMV); Tobravirus: tobacco rattle virus (TRV); Hordeivirus: barley stripe mosaic virus (BSMV); Pecluvirus: Indian peanut clump virus (IPCV), peanut clump virus (PCV).