Family: Alphaflexiviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Distinguishing features
The family contains viruses with flexuous filamentous virions that infect plants and a few viruses discovered in plant-infecting fungi. They share a distinct lineage of alphavirus-like replication proteins that is unusual in lacking any recognized protease domain.
Virion properties
Morphology
Virions are flexuous filaments, usually 12–13 nm in diameter (range 10–15 nm) and from 470 to about 800 nm in length, depending on the genus. They have helical symmetry with a pitch of about 3.4 nm (range 3.3–3.7 nm) and in some genera there is clearly visible cross-banding.
Physicochemical and physical properties
Virions sediment as a single band (or occasionally two very close bands) with an S20,w of 92–176S, depending on the genus.
Nucleic acid
Virions contain a single molecule of linear ssRNA of about 5.9–9.0 kb which is 5–6% by weight of the virion. The RNA is capped (or probably capped) at the 5′ terminus with m7G and has a polyadenylated tract at the 3′ terminus. Smaller 3′-co-terminal sgRNAs are encapsidated in some, but not all, members of the genus Potexvirus.
Proteins
The viral capsid of all members of the family (except in the genus Lolavirus) is composed of a single polypeptide ranging in size from 18 to 43 kDa. In allexiviruses, a 42 kDa polypeptide was also detected as a minor component of virions. In lolaviruses a shorter (ca. 28 kDa) carboxy co-terminal polypeptide forms an equimolar fraction of the virion with the polypeptide originating from the first AUG (ca. 32 kDa).
Lipids
None reported.
Carbohydrates
Usually none, but the coat protein of some strains of the species Potato virus X (genus Potexvirus) and both forms of the lolavirus coat protein are reported to be glycosylated.
Genome organization and replication
There are five or six genes depending upon the genus (except in the genus Sclerodarnavirus). The ORF1-encoded product, which follows a short 5’-UTR sequence, has homologies with polymerase proteins of the “alphavirus-like” supergroup of RNA viruses. This protein (150–195 kDa) contains conserved methyltransferase, helicase and RNA-dependent RNA polymerase (RdRp) motifs; some also include an AlkB domain (alkylated DNA repair protein). In all plant-infecting members, ORFs 2–4 encode the “triple gene block” (TGB) proteins involved in cell-to-cell movement and ORF5 is the viral coat protein. In some genera (Allexivirus, Lolavirus and Mandarivirus) a final ORF encodes a protein with a zinc binding finger motif and the ability to bind nucleic acids. ORFs downstream of the polymerase are translated from 3’-terminal sgRNAs that can often be found in infected tissue. Replication is (or is presumed to be) cytoplasmic and the product of ORF1 is the only virus-encoded protein known to be involved.
Antigenic properties
Virions are usually highly immunogenic. Within (but not usually between) genera, some viruses are serologically related.
Biological properties
Members have been reported from a wide range of mono- and dicotyledonous plant species but the host range of individual members is usually limited. Many of the viruses have relatively mild effects on their host. All species can be transmitted by mechanical inoculation, often readily. Many of the viruses have no known invertebrate or fungus vectors; however, allexiviruses are thought to be mite-borne. Aggregates of virus particles accumulate in the cytoplasm but there are usually no specific cytopathic structures.
Species and genus demarcation criteria in the family
Genera are distinguished by various features of genome organization and host. These are summar-ized in Table 1. Throughout the family, isolates of different species should have less than about 72% nt identity (or 80% aa identity) between their respective CP or polymerase genes. Viruses from different genera usually have less than about 45% nt identity in these genes.
Table 1 Distinguishing properties of genera in the family Alphaflexiviridae
| Genus | Host | Virion length (nm) | ORFs | Repa (kDa) | CPb |
| Allexivirus | plants | ca. 800 | 6 | 170–195 | 26–29 |
| Botrexvirus | fungi | ca.720 | 5 | 158 | 43 |
| Lolavirus | plants | 640 | 6 | 196 | 32 |
| Mandarivirus | plants | 650 | 6 | 187 | 34 |
| Potexvirus | plants | 470–580 | 5 | 150–195 | 18–27 |
| Sclerodarnavirus | fungi | n/ac | 1 | 193 | n/ac |
a Rep, replication protein size (kDa).
b CP, coat protein size (kDa).
c No virions found.
Genus Allexivirus
Type species Shallot virus X
Distinguishing features
Allexiviruses are distinguished by mite transmission and by the presence of a large ORF4 in the position where the third, and smallest, of the TGB proteins is found in the other plant-infecting members of the family.
Virion properties
Morphology
Virions are highly flexible filamentous particles, about 800 nm in length and 12 nm in diameter. They resemble potyviruses in their length, but closteroviruses in their flexibility and cross-banded substructure (Figure 1).
Physicochemical and physical properties
Virions of shallot virus X sediment with an S20,w of about 170S in 0.1 M tris-HC1, pH 7.5 at 20 °C and have a buoyant density in CsCl of 1.33 g cm−3.
Nucleic acid
Virions contain a single molecule of linear ssRNA, about 9.0 kb in size, with a 3’-poly(A) tract.
Proteins
Virions are composed of a 28–36 kDa polypeptide as a major CP. A 42 kDa polypeptide is a minor component of virions.
Genome organization and replication
The genomic RNA contains six large ORFs and short UTRs at the 5′ and 3′ termini (Figure 2). The ORFs code for polypeptides of about 195, 26, 11, 42, 28 and 15 kDa, respectively from 5′ end to 3′ end. The 195 kDa polypeptide is the polymerase. The 26 and 11 kDa proteins are similar to the first two proteins encoded by the TGB of related plant viruses and are probably involved in cell-to-cell movement of the virus. There is a coding sequence for a third small (7–8 kDa) TGB protein but it lacks the initiation AUG-codon. The 42 kDa polypeptide (ORF4) has no significant homology with any known proteins but, in plants infected with an isolate of the type species, it was expressed in relatively large amounts and was shown to be involved in virion assembly. The 28 kDa polypeptide is the CP. In SDS-polyacrylamide gel electrophoresis it migrates as an apparently 32–36 kDa protein, which could be due to its high hydrophilicity. The 15 kDa protein has a zinc binding finger motif and an ability to bind nucleic acids. The function of this polypeptide is not known.
Antigenic properties
Allexivirus particles are good immunogens. Some members of the genus are serologically inter-related. Specific antisera and monoclonal antibodies against pure virus particles as well as antisera against recombinant CPs have been used for differentiation purposes.
Biological properties
Host range
Host range is extremely restricted. Some isolates from shallot, onion, garlic and sand leek have been experimentally transmitted to Chenopodium murale, in which they induced local lesions.
Transmission
Allexiviruses are thought to be mite-borne. Garlic viruses C and D have been shown to be transmitted by the eriophyd mite, Aceria tulipae. All are manually transmissible by sap inoculation to healthy host plants. None could be transmitted by aphids.
Geographical distribution
Allexiviruses are widely distributed and probably occur wherever the host plants are grown.
Cytopathic effects
Most induce no visible or only very mild symptoms in many species, although certain isolates can cause severe damage to crops. In infected tissue allexiviruses can induce formation of granular inclusion bodies and small bundles of flexible particles.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Members of distinct species have less than about 72% nt identity (or 80% aa identity) between their CP or polymerase genes.
- Different reactions with antisera.
List of species in the genus Allexivirus
The available information about the identity of the members in the genus Allexivirus is still fragmentary. They are almost always found in mixed infections of vegetatively propagated species and it is difficult or impossible to isolate and/or separate the viruses.
| Garlic mite-borne filamentous virus |
|
|
| Garlic mite-borne filamentous virus-South Korea |
| (GarMbFV-KO) |
| Garlic virus A |
|
|
| Garlic virus A-Japan | [AB010300 = NC_003375] | (GarV-A-JA) |
| Garlic virus B |
|
|
| Garlic virus B-Japan | [AB010301*] | (GarV-B-JA) |
| Garlic virus C |
|
|
| Garlic virus C-Japan | [AB010302 = NC_003376] | (GarV-C-JA) |
| Garlic virus D |
|
|
| Garlic virus D-Japan | [AB010303*] | (GarV-D-JA) |
| Garlic virus E |
|
|
| Garlic virus E-China:Yuhang | [AJ292230 = NC_004012] | (GarV-E-YH) |
| Garlic virus X |
|
|
| Garlic virus X-Korea | [U89243 = NC_001800] | (GarV-X-KO) |
| Shallot virus X |
|
|
| Shallot virus X-Russia | [M97264 = NC_003795] | (ShVX-RU) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Allexivirus but have not been approved as species
None reported.
Genus Botrexvirus
Type species Botrytis virus X
Distinguishing features
The single member of the genus infects a filamentous fungus. The genome lacks the TGB characteristic of plant-infecting members of the family.
Virion properties
Morphology
Virions are flexuous filaments of 720 nm modal length and about 13 nm in diameter.
Physicochemical and physical properties
No information.
Nucleic acid
Virions contain a single molecule of linear ssRNA, 6966 nt in length, excluding the 3’-poly(A) tail.
Proteins
The only structural protein is the coat protein composed of 400 aa (43 kDa).
Genome organization and replication
The genomic RNA comprises five putative ORFs on the positive strand, a 5′-UTR of 95 nt and a 3’-UTR of 149 nt, followed by a poly(A) tail (Figure 3). ORF1 is the 158 kDa polymerase, ORF3 encodes the 44 kDa coat protein and the functions of the other predicted proteins (ORF2, 30 kDa; ORFs 4 and 5 both 14 kDa) are unknown.
Antigenic properties
No information.
Biological properties
The virus was discovered infecting an isolate of the plant pathogenic fungus Botrytis cinerea. Its mode of transmission is unknown. The same fungal isolate was also infected with a virus now classified as Botrytis virus F (genus Mycoflexivirus, family Gammaflexiviridae).
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Botrexvirus
| Botrytis virus X |
|
|
| Botrytis virus X-New Zealand:Auckland | [AY055762 = NC_005132] | (BotVX-AUK) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Botrexvirus but have not been approved as species
None reported.
Genus Lolavirus
Type species Lolium latent virus
Distinguishing features
This genus consists of a single species. There are probably 6 ORFs (although the putative ORF6 is smaller than in other genera). The coat protein is larger and the virions are longer than the potex-viruses, which it otherwise resembles. Notably two carboxy co-terminal forms of the coat protein are found in essentially equimolar amounts in both extracts of infected plants and in purified virions.
Virion properties
Morphology
Virions are slightly flexuous filaments of 640 nm modal length and about 13 nm diameter (Figure 4).
Physicochemical and physical properties
Virions have a buoyant density in CsCl of about 1.33 g cm−3.
Nucleic acid
Virions contain a single molecule of linear ssRNA, 7674 nt in length, excluding the poly(A) tail. Infectious clones have been produced.
Proteins
The only structural proteins are two carboxy co-terminal forms of the coat protein, composed of 293 amino acids (32 kDa) and about 28 kDa. The two protein forms occur in virions in equimolar amounts. Both forms are glycosylated at one or more of several potential sites.
Genome organization and replication
The genomic RNA is comprised of six putative ORFs on the positive strand, a 5′-UTR of 87 nt and a 3′-UTR of 97 nt, followed by a poly(A) tail (Figure 5). ORF 1 (196 kDa) contains conserved methyl transferase, AlkB, helicase and RdRp motifs. ORFs 2–4 encode the TGB proteins of 30.5, 13 and 7.5 kDa respectively. ORF 5 encodes the viral coat protein of 31.6 kDa; the smaller carboxy co-terminal coat protein is potentially derived by internal initiation at a second AUG 141 nt downstream from the first (and in a stronger context) but might also arise from proteolytic removal of a non-canonical chloroplast transit peptide. ORF 6 partially overlaps ORF 5, and encodes a predicted 45 amino acid, 5.1 kDa highly basic protein (pI 9.56), which may act as a nucleic acid binding protein (NABP); three Cys and two His residues differ in spacing from those of characterized NABPs (10–23 kDa), and no significant homology is observed to other proteins in the database.
Antigenic properties
The virus is a good immunogen and its antiserum reacts with both forms of the coat protein. In indirect ELISA, the antiserum reacted to an isolate of Alternanthera mosaic virus (genus Potexvirus) in infected Nicotiana benthamiana. However, the virus was not detected with antiserum specific to this or other viruses in the genera Potexvirus and Carlavirus.
Biological properties
Host range
The natural host range of the virus is restricted to gramineaceous species including ryegrass (Lolium). It also readily infects N. benthamiana and a few other dicotyledonous species. In N. benthamiana, it induces local lesions and systemic mosaic that varies from mildly chlorotic to white.
Transmission
Readily transmitted to healthy host plants by sap inoculation.
Geographical distribution
Reported from Europe (Germany, the Netherlands, France and the United Kingdom), and from the United States.
Cytopathic effects
The virus induces insignificant symptoms or mild chlorotic flecking in its natural hosts. More severe chlorotic to necrotic streaking may occur in mixed infectious with other viruses. In infected tissue masses of flexuous virions may be observed in association with the chloroplasts.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Lolavirus
| Lolium latent virus |
|
|
| Lolium latent virus-US1 | [EU489641 = NC_010434] | (LoLV-US1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Lolavirus but have not been approved as species
None reported.
Genus Mandarivirus
Type species Indian citrus ringspot virus
Distinguishing features
This genus contains a single virus species infecting a tree host. There are six ORFs in the genome and the coat protein is the largest of any plant-infecting member of the family.
Virion properties
Morphology
Virions are flexuous filaments of 650 nm modal length, 13 nm in diameter, with clearly visible cross-banding (Figure 6).
Physicochemical and physical properties
The virus forms a single band in caesium sulphate density gradients. Purified preparations show maximum absorption at 260 nm with a A260/280 ratio of 1.1 (corrected for light scattering).
Nucleic acid
Virions contain a single molecule of linear ssRNA, 7560 nt in length, excluding the 3′-poly(A) tail.
Proteins
The only structural protein is the CP, composed of 325 aa (34 kDa).
Genome organization and replication
The genomic RNA comprises six ORFs on the positive strand, a 5′-UTR of 78 nt and a 3′-UTR of 40 nt, followed by a poly(A) tail (Figure 7). No significant ORFs are in the negative strand. ORF1 encodes the viral polymerase. ORFs 2, 3 and 4 form the TGB. ORF5 encodes the CP. ORF6 encodes a putative protein of unknown function that shows limited similarity with nucleic acid-binding proteins encoded by ORF6 of allexi- and carlaviruses.
Antigenic properties
Particles are good immunogens, rabbit antisera can have titers of 1/128 and 1/2048 in gel diffusion and EM decoration, respectively.
Biological properties
The virus causes a serious disease of citrus, especially Kinnow mandarin, in India, with bright yellow ringspots on mature leaves, followed by rapid decline of the tree. Experimentally the virus can be mechanically inoculated to leaves of Chenopodium quinoa, C. amaranticolor, Glycine max, Vigna unguiculata and Phaseolus vulgaris, giving local lesions, but systemic infection only in P. vulgaris. No natural vector is known, but it is transmitted by grafting and persists in the host.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Mandarivirus
| Indian citrus ringspot virus |
|
|
| Indian citrus ringspot virus-K1 | [AF406744 = NC_003093] | (ICRSV-K1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Mandarivirus but have not been approved as species
None reported.
Genus Potexvirus
Type species Potato virus X
Distinguishing features
Compared to other genera in the family, potexviruses have short virions (<700 nm) and only five ORFs. They infect herbaceous hosts and have no known vectors.
Virion properties
Morphology
Virions are flexuous filaments, 470–580 nm in length and 13 nm in diameter, with helical symmetry and a pitch of 3.3–3.7 nm (Figure 8). A central axial canal, about 3 nm in diameter, is discernible only in best preparations. The number of protein subunits per turn of the primary helix is slightly less than 9.0. The RNA backbone is at a radial position of 3.3 nm.
Physicochemical and physical properties
Virion Mr is about. 3.5×106; S20,w is 115–130S; buoyant density in CsCl is 1.31 g cm−3.
Nucleic acid
Virions contain a single linear molecule of positive sense ssRNA of about. 5.9–7.0 kb which is approximately 6% by weight of the virion. The RNA is capped at the 5′ terminus with m7G and has a polyadenylated tract at the 3′ terminus.
Proteins
The virus capsid consists of 1000–1500 protein subunits of a single 18–27 kDa polypeptide. Partial proteolytic cleavage of the CP subunits can occur during storage of purified virus.
Genome organization and replication
Virions of the type member contain only genomic RNA, but other potexviruses also encapsidate the sgRNA for the CP. Genomic RNA is translated as a functionally monocistronic message; only the 5′-proximal RNA-polymerase gene is translated directly by ribosomes, producing the RNA polymerase (150–181 kDa). The 5′-UTR leader sequence of PVX RNA consists of 83 nt (excluding the cap-structure) and efficiently enhances translation. In infected plants, some potexviruses produce sgRNAs including one that acts as messenger RNA for the CP (Figure 9).
The genomic RNA of potexviruses typically has five ORFs. ORF1, at the 5′ terminus, is the polymerase gene and ORF5, located at the 3′ terminus, is the CP gene. Between ORF1 and ORF5 is the TGB of three overlapping ORFs, the products of which (25, 12 and 8 kDa) are involved in cell-to-cell movement of viral RNA. The 25 kDa protein (as well as the 166 kDa replicase) contains an NTPase-helicase domain, but is not involved in RNA replication. It has been shown to have RNA silencing suppressor activity which is necessary for virus movement. The 12 and 8 kDa proteins contain large blocks of uncharged aa and are associated with membrane vesicles derived from the endoplasmic reticulum. TGB3 of AltMV (but not that of PVX) is targeted to the chloroplast and is required for movement from the epidermis to the mesophyll layer. The CP is also involved in cell-to-cell movement. ORFs 2 to 5 are expressed via the production (and subsequent translation) of appropriate sgRNAs. Two or three 3′-co-terminal sgRNAs can be isolated from plants infected with potexviruses (2.1, 1.2 and 1.0 kb); the double stranded counterparts of these sgRNAs have also been detected. The medium-size sgRNA (1.2 kb) is probably functionally bicistronic, its translation yielding the 12 and 8 kDa proteins.
Antigenic properties
Virions are highly immunogenic; some species are antigenically related, but others are serologically distinct.
Biological properties
Host range
Some of the viruses are moderately pathogenic, causing mosaic or ringspot symptoms in a wide range of mono- and dicotyledonous plant species, but others alone cause little damage to infected plants. The host range of individual members is limited.
Transmission
The viruses are transmitted in nature by mechanical contact. Potato aucuba mosaic virus can be vectored by aphids when assisted by a potyvirus providing a helper protein.
Geographical distribution
As a group, potexviruses occur world-wide. The distribution of some species is very wide but others are apparently more restricted.
Cytopathic effects
The cytoplasm of infected cells contains fibrous, banded or irregular aggregates of virus particles, and often membrane accumulations. There is no cytopathology specific to potexviruses, although some viruses induce unique structures such as the beaded sheets found in cells infected by the type member.
Species demarcation criteria in the genus
The list of species demarcation criteria in the genus is:
- Host range: the natural host range is usually specific to different species.
- Distinct species fail to cross-protect in infected plants.
- Serology: species and strains of some species are also readily distinguishable in differential reactions with monoclonal antibodies.
- Sequence: isolates of different species have less than about 72% nt identity (or 80% aa identity) between their CP or polymerase genes.
List of species in the genus Potexvirus
| Alstroemeria virus X |
|
|
| Alstroemeria virus X-Japan | [AB206396 = NC_007408] | (AlsVX-JA) |
| Alternanthera mosaic virus |
|
|
| Alternanthera mosaic virus-USA:Pennsylvania | [AY863024 = NC_007731] | (AltMV-PA) |
| Asparagus virus 3 |
|
|
| Asparagus virus 3-Japan | [AB304848 = NC_010416] | (AV3-J) |
| Scallion virus X-China: Hangzhou | [AJ316085 = NC_003400] | (ScaVX-HZ) |
| Bamboo mosaic virus |
|
|
| Bamboo mosaic virus-O | [D26017 = NC_001642] | (BaMV-O) |
| Cactus virus X |
|
|
| Cactus virus X-Taiwan | [AF308158 = NC_002815] | (CVX-TW) |
| Cassava common mosaic virus |
|
|
| Cassava common mosaic virus-Brazil | [U23414 = NC_001658] | (CsCMV-BR) |
| Cassava virus X |
|
|
| Cassava virus X-Colombia |
| (CsVX-COL) |
| Clover yellow mosaic virus |
|
|
| Clover yellow mosaic virus-USA | [D29630 = NC_001753] | (ClYMV-USA) |
| Commelina virus X |
|
|
| Commelina virus X-United Kingdom |
| (ComVX-UK) |
| Cymbidium mosaic virus |
|
|
| Cymbidium mosaic virus-Singapore | [U62963 = NC_001812] | (CymMV-SIN) |
| Daphne virus X |
|
|
| Daphne virus X-New Zealand |
| (DVX-NZ) |
| Foxtail mosaic virus |
|
|
| Foxtail mosaic virus-USA | [M62730 = NC_001483] | (FoMV-USA) |
| Hosta virus X |
|
|
| Hosta virus X-type strain: HVX-Kr | [AJ620114 = NC_011544] | (HVX-Kr) |
| Hydrangea ringspot virus |
|
|
| Hydrangea ringspot virus-PD 109 | [AY707100 = NC_006943] | (HdRSV-PD109) |
| Lettuce virus X |
|
|
| Lettuce virus X-Iran:Karaj | [AM745758 = NC_010832] | (LeVX-IR) |
| Lily virus X |
|
|
| Lily virus X-Netherlands | [AJ633822 = NC_007192] | (LVX-NL) |
| Malva mosaic virus |
|
|
| Malva mosaic virus-Chenopodium mosaic virus X | [DQ660333 = NC_008251] | (MalMV-ChMVX) |
| Mint virus X |
|
|
| Mint virus X-USA | [AY789138 = NC_006948] | (MVX-USA) |
| Narcissus mosaic virus |
|
|
| Narcissus mosaic virus-Netherlands | [D13747 = NC_001441] | (NMV-NL) |
| Nerine virus X |
|
|
| Nerine virus X-Japan | [AB219105 = NC_007679] | (NVX-J) |
| Opuntia virus X |
|
|
| Opuntia virus X-CC10 | [AY366209 = NC_006060] | (OpVX-CC10) |
| Papaya mosaic virus |
|
|
| Papaya mosaic virus-USA:Florida | [D13957 = NC_001748] | (PapMV-FLA) |
| Pepino mosaic virus |
|
|
| Pepino mosaic virus-Sp-13 | [AF484251 = NC_004067] | (PepMV-Sp13) |
| Phaius virus X |
|
|
| Phaius virus X-Japan | [AB353071 = NC_010295] | (PhVX-JA) |
| Plantago asiatica mosaic virus |
|
|
| Plantago asiatica mosaic virus-Russia | [Z21647 = NC_003849] | (PlAMV-RU) |
| Nandina mosaic virus-USA | [AY800279] | (NaMV-USA) |
| Plantago severe mottle virus |
|
|
| Plantago severe mottle virus-Canada |
| (PlSMoV-CN) |
| Plantain virus X |
|
|
| Plantain virus X-United Kingdom |
| (PlVX-UK) |
| Potato aucuba mosaic virus |
|
|
| Potato aucuba mosaic virus-Netherlands | [S73580 = NC_003632] | (PAMV-NL) |
| Potato virus X |
|
|
| Potato virus X-X3 | [D00344 = NC_011620] | (PVX-X3) |
| Schlumbergera virus X |
|
|
| Schlumbergera virus X-K11 | [AY366207 = NC_011659] | (SchVX-K11) |
| Strawberry mild yellow edge virus |
|
|
| Strawberry mild yellow edge virus-MY-18 | [D12517 = NC_003794] | (SMYEV-MY18) |
| Tamus red mosaic virus |
|
|
| Tamus red mosaic virus-Italy |
| (TRMV-IT) |
| Tulip virus X |
|
|
| Tulip virus X-J | [AB066288 = NC_004322] | (TVX-J) |
| White clover mosaic virus |
|
|
| White clover mosaic virus-USA | [X06728 = NC_003820] | (WClMV-USA) |
| Zygocactus virus X |
|
|
| Zygocactus virus X-B1 | [AY366208 = NC_006059] | (ZyVX-B1) |
Species names are in italic script; names of strains and isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Potexvirus but have not been approved as species
| Allium virus X | [FJ670570 = NC_012211] | (AlVX) |
| Caladium virus X | [AY727533*] | (CalVX) |
| Dioscorea latent virus |
| (DLV) |
| Paris polyphylla virus X | [DQ530433*] | (PPVX) |
| Parsnip virus 3 |
| (ParV-3) |
| Viola mottle virus |
| (VMoV) |
| Wineberry latent virus |
| (WLV) |
* Sequences do not comprise the complete genome.
Genus Sclerodarnavirus
Type species Sclerotinia sclerotiorum debilitation-associated RNA virus
Distinguishing features
The genus consists of a single member, a capsid-less mycovirus. Despite the lack of capsid, phylogenetic analysis of the polymerase places it within this family.
Virion properties
Morphology
There are no virions.
Physicochemical and physical properties
No information.
Nucleic acid
A single linear molecule of positive sense ssRNA of 5470 nt with a polyadenylated tract at the 3′ terminus.
Genome organization and replication
There is a single ORF encoding a replication protein of 193 kDa.
Antigenic properties
Not applicable.
Biological properties
The single member was discovered in the plant pathogenic fungus Sclerotinia sclerotiorum and appears to cause debilitation (hypovirulence).
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Sclerodarnavirus
| Sclerotinia sclerotiorum debilitation-associated RNA virus |
|
|
| Sclerotinia sclerotiorum debilitation-associated RNA virus-China | [AY147260 = NC_007415] | (SSDaRV-CN) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Sclerodarnavirus but have not been approved as species
None reported.
List of unassigned species in the family Alphaflexiviridae
None.
List of other related viruses which may be members of the family Alphaflexiviridae but have not been approved as species
| Ambrosia asymptomatic virus 1 | [EU362849-50*] | (ASV1) |
| Blackberry virus X | [DQ378057*] | (BlVX) |
* Sequences do not comprise the complete genome.
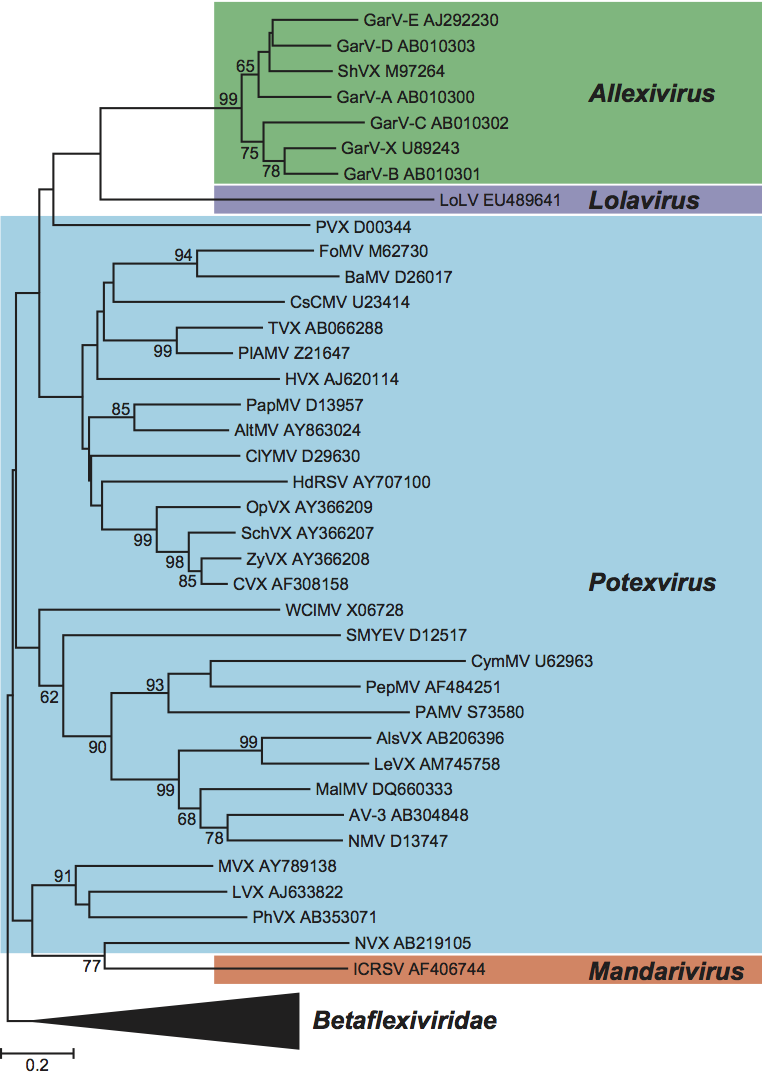
Phylogenetic relationships within the family
In a phylogenetic analysis of the replication protein, most genera fall on well-supported branches but the single member of the genus Mandarivirus groups with Potexvirus (Figure 10, p. 917). Similar results are obtained in analyses of the first (and largest) TGB protein (Figure 11) although members of the genus Potexvirus do not form such a coherent group.
Similarity with other taxa
The polymerase proteins are members of the “alphavirus-like” supergroup of RNA viruses and are most closely related to those of the other families in the order, namely Betaflexiviridae, Gammaflexiviridae and Tymoviridae. The TGB proteins are related to those of some genera in the family Betaflexiviridae and, more distantly, to those of rod-shaped viruses in the family Virgaviridae (genera Hordeivirus, Pecluvirus and Pomovirus).
Derivation of names
Allexi: from Allium (the genus name for the principal host, shallot) + X.
Botrex: from Botrytis virus X.
Flexi: from Latin flexus, “bent”.
Lola: from Lolium latent virus.
Mandari: from mandarin (Citrus reticulata), the host of the type species, Indian citrus ringspot virus.
Potex: from Potato virus X.
Sclerodarna: from Sclerotinia sclerotiorum debilitation-associated RNA virus.
Further reading
Adams et al., 2004 M.J. Adams, J.F. Antoniw, M. Bar-Joseph, A.A. Brunt, T. Candresse, G.D. Foster, G.P. Martelli, R.G. Milne, S.K. Zavriev, C.M. Fauquet, The new plant virus family Flexiviridae and assessment of molecular criteria for species demarcation. Arch. Virol. 149 (2004) 1045–1060.
Martelli et al., 2007 G. Martelli, M.J. Adams, J.F. Kreuze, V.V. Dolja, Family Flexiviridae: a case study in virion and genome plasticity. Annu. Rev. Phytopathol. 45 (2007) 73–100.
Verchot-Lubicz et al., 2007 J. Verchot-Lubicz, C.-M. Ye, D. Bamunusinghe, Molecular biology of potexviruses: Recent advances. J. Gen. Virol. 88 (2007) 1643–1655.
Contributed by
Adams, M.J., Candresse, T., Hammond, J., Kreuze, J.F., Martelli, G.P., Namba, S., Pearson, M.N., Ryu, K.H. and Vaira, A.M.
Figures
Figure 1 Electron micrograph of negatively-stained virions of an isolate of shallot virus X. The bar = 200 nm.
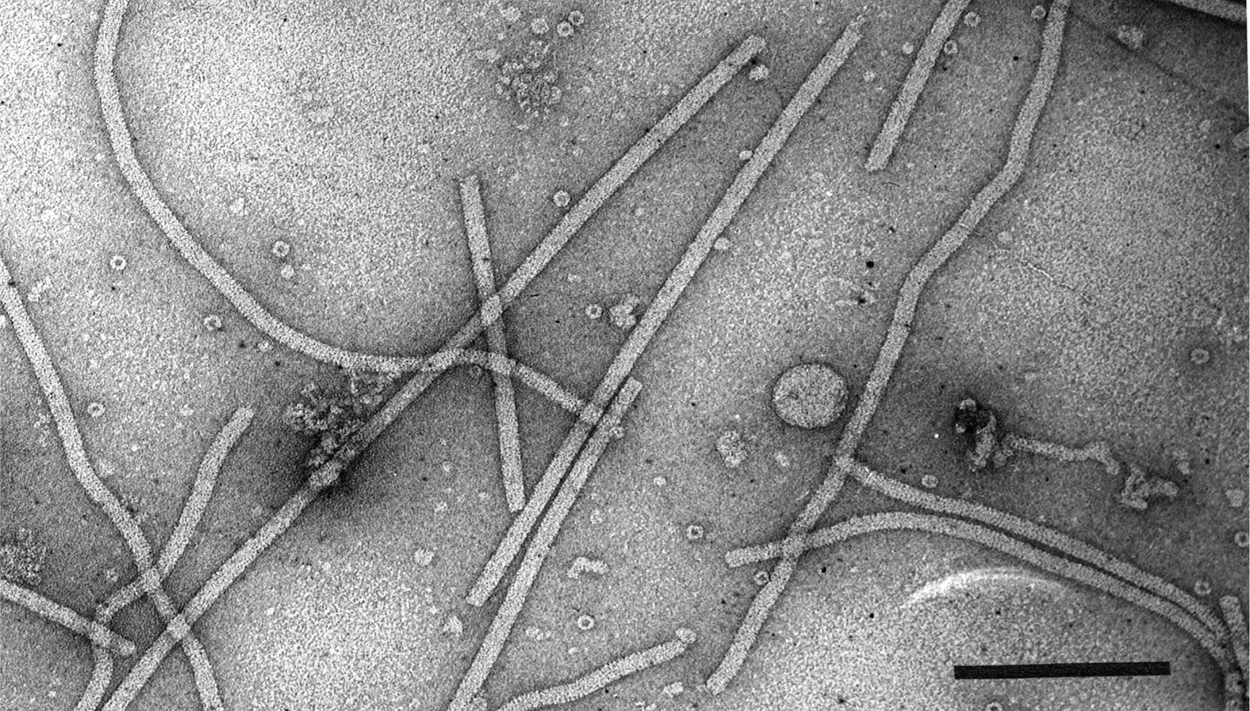
Figure 2 Genome organization and translation strategy of shallot virus X.
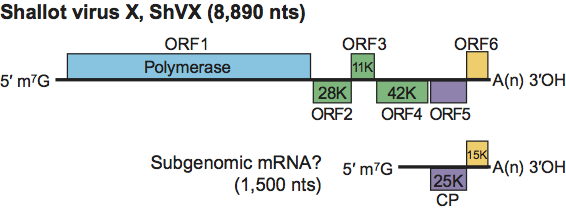
Figure 3 Genome organization of Botrytis virus X showing the relative positions of the ORFs and their expression products. Mtr, methyltransferase; Hel, helicase; RdRp, RNA-dependent RNA polymerase; CP, capsid protein.
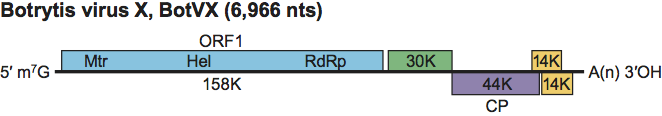
Figure 4 Negative contrast electron micrograph of particles of an isolate of Lolium latent virus stained in phosphotungstic acid. The bar represents 500 nm.
(Courtesy of M.M. Dienelt.)
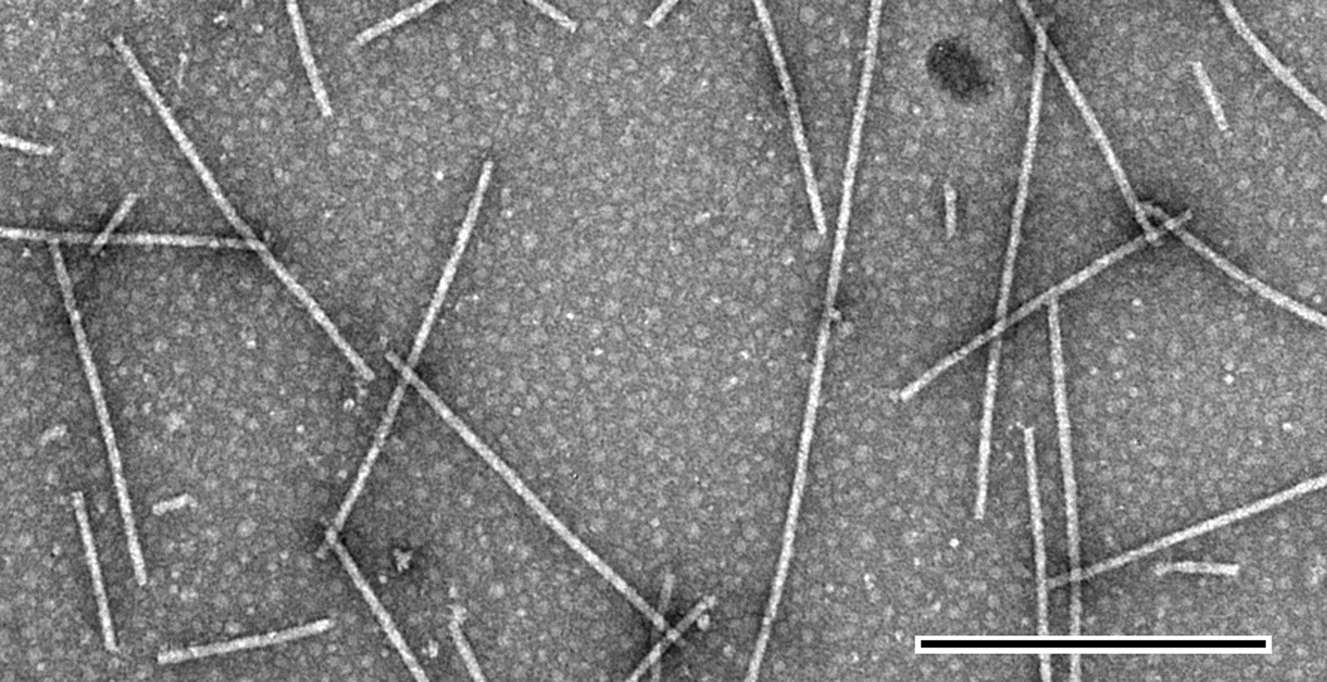
Figure 5 Genome organization of Lolium latent virus showing the relative positions of the ORFs and their expression products. Mtr, methyltransferase; Hel, helicase; RdRp, RNA-dependent RNA polymerase; TGB, triple gene block; CP, capsid protein; NB, putative nucleic acid binding protein.
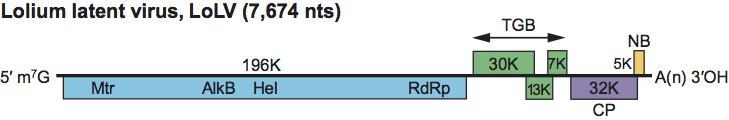
Figure 6 Electron micrograph of particles of an isolate of Indian citrus ringspot virus, stained in 1% uranyl acetate. The bar represents 100 nm.
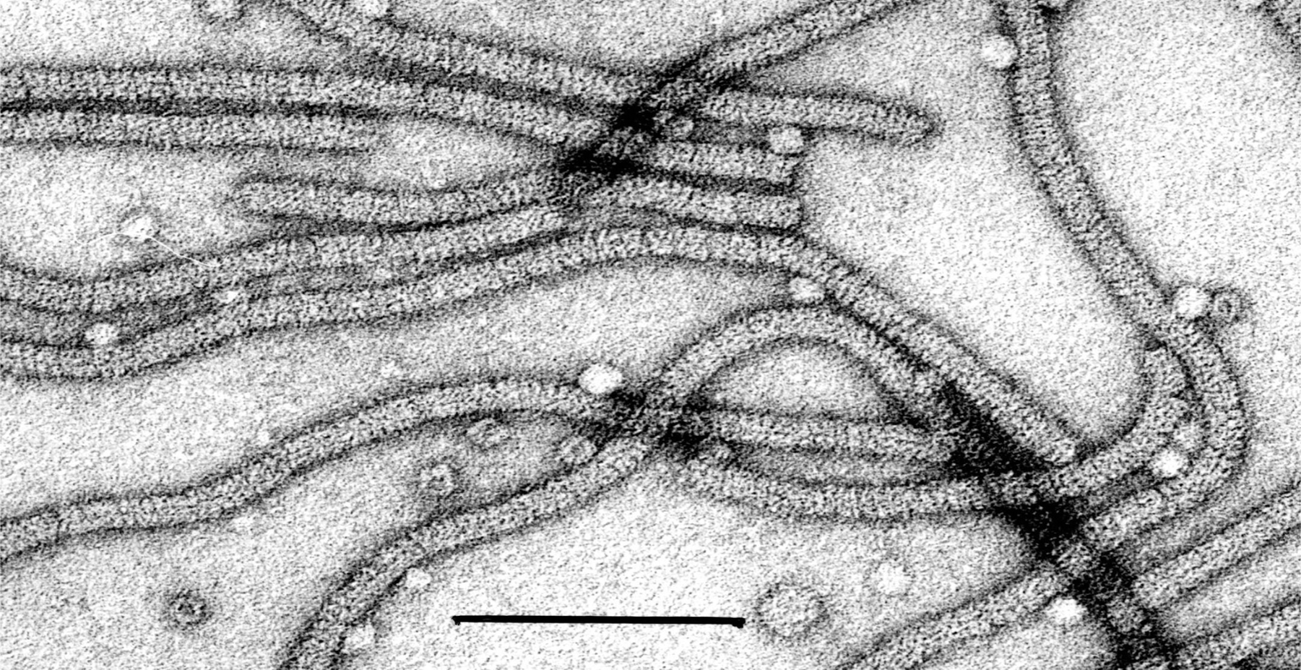
Figure 7 Genome organization of Indian citrus ringspot virus (ICRSV) showing the relative positions of the ORFs and their expression products. Mtr, methyltransferase; Hel, helicase; RdRp, RNA-dependent RNA polymerase; CP, capsid protein; NB, nucleic acid binding protein. The 25K, 12K and 6.4K proteins constitute the triple gene block.
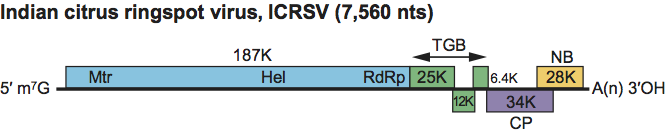
Figure 8 Negative contrast electron micrograph of particles of an isolate of potato virus X. The bar represents 100 nm.
(Courtesy of D.-E. Lesemann.)
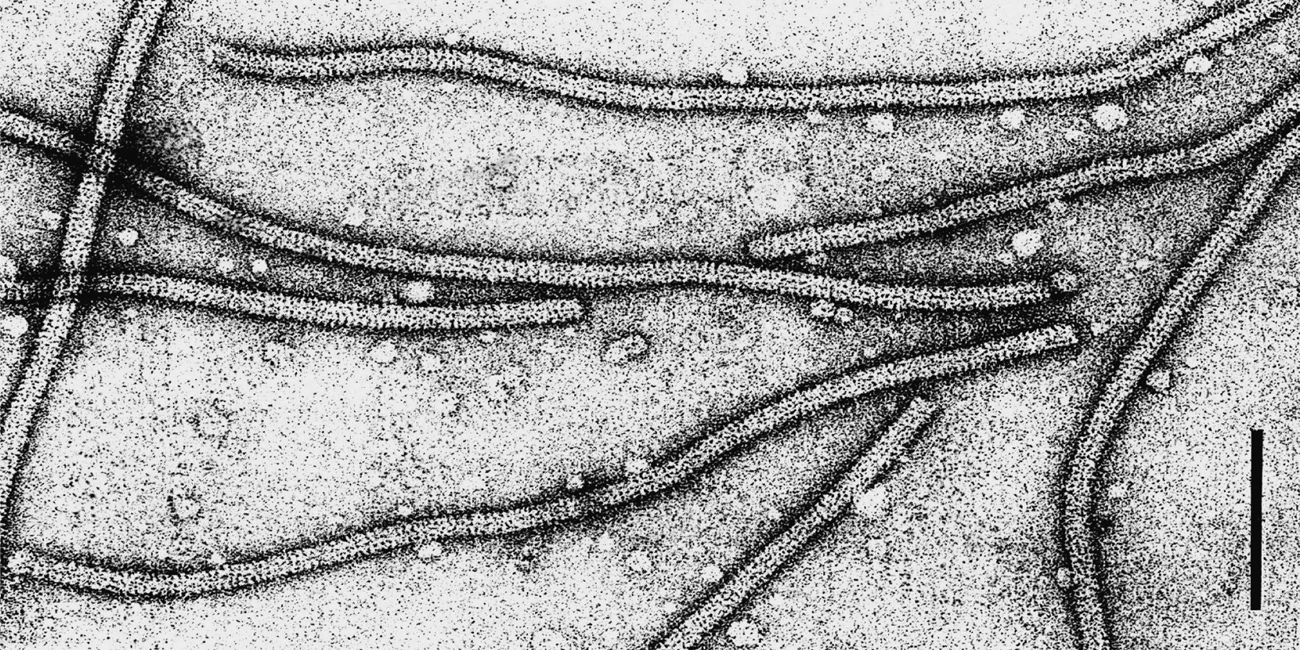
Figure 9 Potato virus X genome organization and expression.
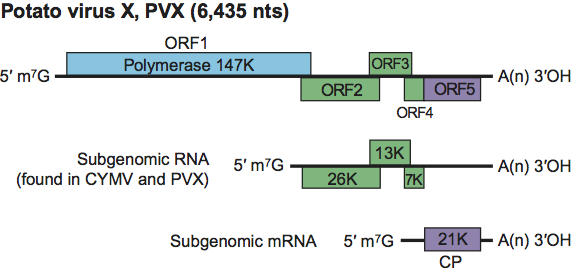
Figure 10 Phylogenetic (distance) tree based on the amino acid sequences of the entire replication protein of members of the family Alphaflexiviridae. A single representative isolate of each sequenced species in the family was included and the tree is rooted with Botrytis virus F (BotV-F; genus Mycoflexivirus, family Gammaflexiviridae). Numbers on branches indicate percentage of bootstrap support out of 1000 bootstrap replications (when >60%). The scale indicates JTT amino acid distances. Tree produced in MEGA4.
Figure 11 Phylogenetic (distance) tree based on the amino acid sequences of the first triple gene block protein of members of the family Alphaflexiviridae. A single representative isolate of each sequenced species in the family was included; the tree also contains similar sequences from the family Betaflexiviridae collapsed into a triangle, the length of which corresponds to the variation found within the clade. Numbers on branches indicate percentage of bootstrap support out of 1000 bootstrap replications (when >60%). The scale indicates JTT amino acid distances. Tree produced in MEGA4.