Genus: Tenuivirus
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
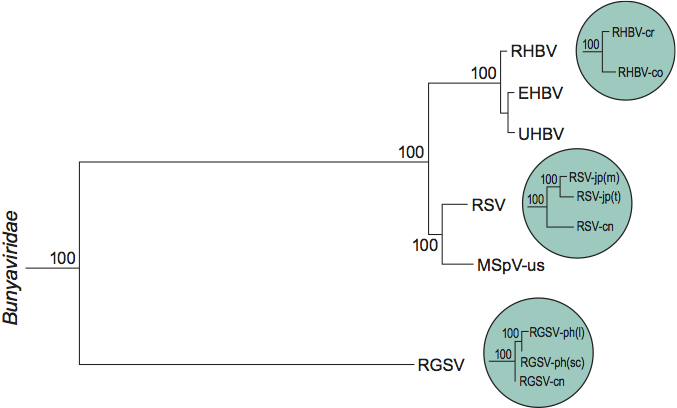
The ribonucleoproteins (RNPs) have a thin filamentous shape; they consist of nucleocapsids, 3–10 nm in diameter, with lengths proportional to the sizes of the RNAs they contain. The filamentous particles may appear to be spiral-shaped, branched or circular (Figure 1). No envelope has been observed.
Physicochemical and physical properties
RNP preparations can be separated into four or five components by sucrose density gradient centrifugation, but form one component with a buoyant density 1.282–1.288 g cm−3 when centrifuged to equilibrium in CsCl solutions.
Nucleic acid
The ssRNA genome consists of four or more segments. The sizes are about 9 kb (RNA-1, generally of negative polarity), 3.3–3.6 kb (RNA-2, ambisense), 2.2–2.5 kb (RNA-3, ambisense), and 1.9–2.2 kb (RNA-4, ambisense). RNP preparations of maize stripe virus (MSpV) and Echinochloa hoja blanca virus (EHBV) contain a fifth RNA of negative polarity and with a size of 1.3 kb. A fifth RNA segment has also been reported for some isolates of rice stripe virus (RSV). Rice grassy stunt virus (RGSV) preparations contain six segments, all of which are ambisense. RGSV RNAs-1, -2, -5 and -6 are homologous to RNA-1, -2, -3 and -4 respectively of other tenuiviruses, whereas RNA-3 (3.1 kb) and RNA-4 (2.9 kb) are unique to RGSV.
Proteins
The nucleocapsid proteins are of 34–35 kDa. Small amounts of a minor 230 kDa protein co-purify with RNPs of RSV, rice hoja blanca virus (RHBV) and RGSV. This protein may be an RdRp polymerase, as this activity is associated with filamentous nucleoprotein particles.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The 3′- and 5′-terminal sequences of each ssRNA are almost complementary for about 20 bases. Several RNA segments encode two proteins in an ambisense arrangement (Figure 2). In most tenuiviruses, the nucleocapsid protein (pC3; N) is encoded by the 5′-proximal region of the virion-complementary sense strand of RNA-3. Virion-sense RNA-4 encodes in its 5′-proximal region a major non-structural protein (p4; NCP) that accumulates in infected plants. Some of the intergenic non-coding regions (NCRs) between the ORFs can adopt hairpin structures. Some segments (e.g. RNA-1 of RSV and RNA-5 of MSpV) are of negative polarity. RNA-1 encodes the RdRp (pC1; RdRp). Some proteins are translated from sgRNAs (Figure 3). For MSpV, RHBV and RSV mRNAs, the production of mRNAs involves a cap-snatching mechanism. An RNA polymerase has been found associated with purified preparations of RSV, RHBV and RGSV. The RNA polymerase activity of RHBV is capable of replicating and transcribing the RNA segments in vitro. In RHBV and RSV, p3 is a suppressor of RNA silencing. In RSV, pC4 is a movement protein.
Antigenic properties
The N proteins of RSV and MSpV are serologically related, and both the N and NCP proteins of RSV and RGSV are related. Likewise, the N proteins of RHBV, EHBV and Urochloa hoja blanca virus (UHBV) are serologically related. The RSV N protein reacts weakly with antibodies made to virion preparations of RGSV or RHBV.
Biological properties
Host range
Plant hosts of tenuiviruses are all in the family Gramineae.
Transmission
Each species is transmitted by a particular species of planthopper in a circulative, propagative manner. The major vectors are Laodelphax striatellus (RSV), Peregrinus maidis (MSpV), Tagosodes orizicolus (RHBV), T. cubanus (EHBV), Nilaparvata lugens (RGSV), Caenodelphax teapae (UHBV), Ukanodes tanasijevici (Iranian wheat stripe virus; IWSV), Javesella pellucida (European wheat striate mosaic virus; EWSMV) and Sogatella kolophon (Brazilian wheat spike virus; BWSV).
Tenuiviruses can be transmitted transovarially by viruliferous female planthoppers to their offspring, and through sperm from viruliferous males. Mechanical transmission using sap extracts is difficult.
Cytopathic effects
Characteristic inclusion bodies, consisting almost entirely of NCP are formed in cells of infected plants. The protein p5 of RGSV accumulates in large amounts in both infected plants and vector insects.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Vector specificity, i.e. transmission by different species of vector
- Host range, i.e. different abilities to infect key plant species
- Different sizes and/or numbers of RNA components
- <85% aa sequence identity between any corresponding gene products
- <60% nt sequence identity between corresponding non-coding intergenic regions
An example of species discrimination is that between RSV and MSpV. RSV is transmitted by L. striatellus and infects 37 species in the Gramineae including wheat and rice. MSpV is transmitted by P. maidis and infects maize, occasionally sorghum and a few other graminaceous plants but not wheat or rice. RSV isolates have genomes of four RNA segments of 9090, 3514, 2475 to 2504 and 2137 to 2157 nt; the MSpV genome has five segments (ca. 9000, 3575, 2357, 2227 and 1317 nt). Also, the differences in sequence among the components of these viruses all fall outside the limits set by the Species Demarcation Criteria.
It is difficult to decide if RHBV, EHBV and UHBV are the same or different species. They have different vectors, different hosts, different sizes and numbers of RNA segments and the nt identity of their intergenic regions is less than 60%. However, the aa sequences of the four proteins on RNA-3 and RNA-4 are about 90% identical between RHBV, EHBV and UHBV (although the nt identities of these same coding regions are about 81% identical among them). So four out of five criteria are met, and therefore they could be considered distinct species, possibly only recently separated and now diverging, with little contact in the field between them.
List of species in the genus Tenuivirus
| Echinochloa hoja blanca virus |
|
|
| Echinochloa hoja blanca virus - cr | [RNA3, L75930; RNA4, L48441; RNA5, L47430] | (EHBV-cr) |
| Maize stripe virus |
|
|
| Maize stripe virus - us | [RNA2, U53224; RNA3, M57426; RNA4, L13438; RNA5, L13446] | (MSpV-us) |
| Rice grassy stunt virus |
|
|
| Rice grassy stunt virus - ph(Laguna) | [RNA1, AB009656; RNA2, AB010376; RNA3, AB010377, RNA4, AB010378; RNA5, AB000403; RNA6, AB000404] | (RGSV-phl) |
| Rice hoja blanca virus |
|
|
| Rice hoja blanca virus - cr | [RNA1, AF009569*; RNA2, L54073; RNA3, L07940; RNA4, AF004657] | (RHBV-cr) |
| Rice stripe virus |
|
|
| Rice stripe virus - jp (t) | [RNA1, D31879; RNA2, D13176; RNA3, X53563; RNA4, D10979] | (RSV-jpt) |
| Urochloa hoja blanca virus |
|
|
| Urochloa hoja blanca virus - cr | [RNA1, U82448*; RNA3, U82447; RNA4, U82446] | (UHBV-cr) |
Species names are in italic script; names of isolates, are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome segment.
List of other related viruses which may be members of the genus Tenuivirus but have not been approved as species
| Brazilian wheat spike virus |
| (BWSpV) |
| European wheat striate mosaic virus |
| (EWSMV) |
| Iranian wheat stripe virus | [RNA2, AY312434; RNA3, AY312435; RNA4, AY312436] | (IWSV) |
| Maize yellow stripe virus | [RNA1, AJ969412*; RNA2, AJ696413*; RNA3, AJ969414*; RNA4, AJ969415*; RNA5, AJ969416] | (MYSV) |
| Rice wilted stunt virus |
| (RWSV) |
| Winter wheat mosaic virus |
| (WWMV) |
* Sequences do not comprise the complete genome segment.
Phylogenetic relationships within the genus
Phylogenetic relationships within the genus are illustrated in Figure 4.
Similarity with other taxa
Tenuiviruses have several similarities with viruses classified in the family Bunyaviridae, particularly those in the genus Phlebovirus. The multipartite genomes of tenuiviruses contain negative sense and ambisense components. RNPs containing the genomic RNAs can be purified from infected plants. The genomic RNA 5′ and 3′ ends can base-pair, and probably give rise to circular RNPs. Generation of mRNA involves a cap-snatching mechanism. Like viruses in most genera in the family Bunyaviridae, tenuiviruses infect their insect vectors as well as their primary hosts, plants. The number of genome components (four to six) and the apparent lack of a membrane-bound virus particle distinguish tenuiviruses from viruses in the family Bunyaviridae. Recent data raise the possibility that RGSV be classified in a separate genus. It has six RNA segments that in total encode four or five proteins in addition to those characteristic of the expression of a tenuivirus genome. Moreover, the sequence relatedness of the RGSV gene products with those of other tenuiviruses are all unusually low. Viruses of the recently established genus Emaravirus also have properties similar to tenuiviruses and members of the Bunyaviridae.
Derivation of names
Tenui: from Latin tenuis, “thin, fine, weak”.
Further reading
Bucher et al., 2003 E. Bucher, T. Sijen, P. de Haan, R. Goldbach, M. Prins, Negative-strand tospoviruses and tenuiviruses carry a gene for a suppressor of gene silencing at analogous genomic positions. J. Virol. 77 (2003) 1329–1336.
de Miranda et al., 2001 J.R. de Miranda, M. Munoz, R. Wu, A.M. Espinoza, Phylogenetic position of a novel tenuivirus from the grass Urochloa plantaginea. Virus Genes. 22 (2001) 329–333.
Falk and Tsai, 1998 B.W. Falk, J.H. Tsai, Biology and molecular biology of viruses in the genus Tenuivirus. Annu. Rev. Phytopathol. 36 (1998) 139–163.
Madriz et al., 1998 J. Madriz, J.R. de Miranda, E. Cabezas, M. Oliva, M. Hernandez, A.M. Espinoza, Echinochloa hoja blanca virus and rice hoja blanca virus occupy distinct ecological niches. J. Phytopath. 146 (1998) 305–308.
Miranda et al., 2000 G.J. Miranda, O. Azzam, Y. Shirako, Comparison of nucleotide sequences between northern and southern philippine isolates of rice grassy stunt virus indicates occurrence of natural genetic reassortment. Virology. 266 (2000) 26–32.
Ramirez and Haenni, 1994 B.-C. Ramirez, A.-L. Haenni, Molecular biology of tenuiviruses, a remarkable group of plant viruses. J. Gen. Virol. 75 (1994) 467–475.
Toriyama et al., 1998 S. Toriyama, T. Kimishima, M. Takahashi, T. Shimizu, N. Minaka, K. Akutsu, The complete nucleotide sequence of the rice grassy stunt virus genome and genomic comparisons with viruses of the genus Tenuivirus. J. Gen. Virol. 79 (1998) 2051–2058.
Xiong et al., 2008 R. Xiong, J. Wu, Y. Zhou, X. Zhou, Identification of a movement protein of the tenuivirus rice stripe virus. J. Virol. 82 (2008) 12304–12311.
Contributed by
Shirako, Y., Falk, B.W. and Haenni, A.-L.
Figures
Figure 1 Electron micrographs of sucrose density gradient purified RNPs of rice hoja blanca virus (RHBV). (Left) Small circular RNPs from the slowest sedimenting RHBV RNP. (Right) Larger, circular RNPs from the fastest sedimenting RHBV RNP. The bar represents 100 nm.
(Courtesy of A.M. Espinoza.)
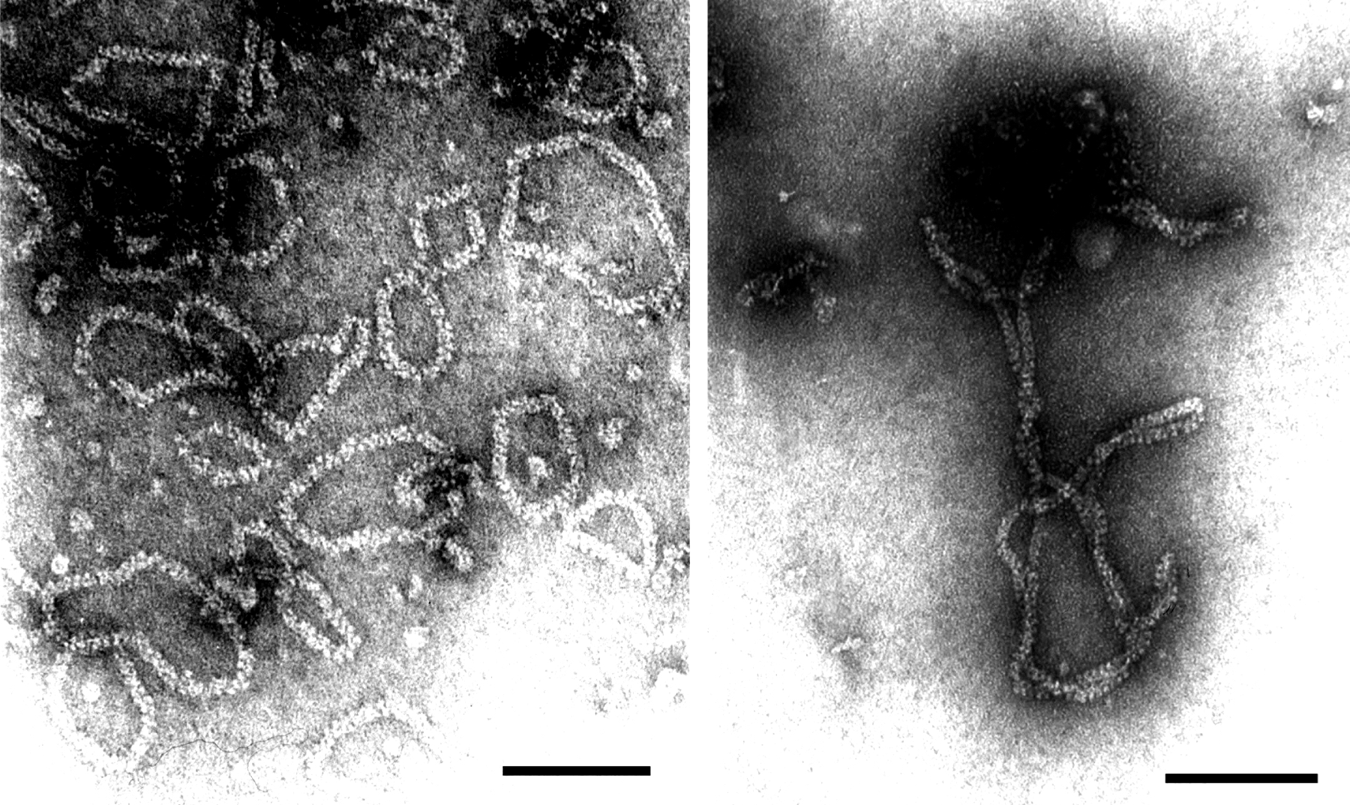
Figure 2 Genome organization characteristic of (top) maize stripe virus (MSpV) and (bottom) rice grassy stunt virus (RGSV). Boxes indicate the positions and designations of the ORF translation products. V signifies virion-sense RNA and VC signifies virion-complementary sense RNA.
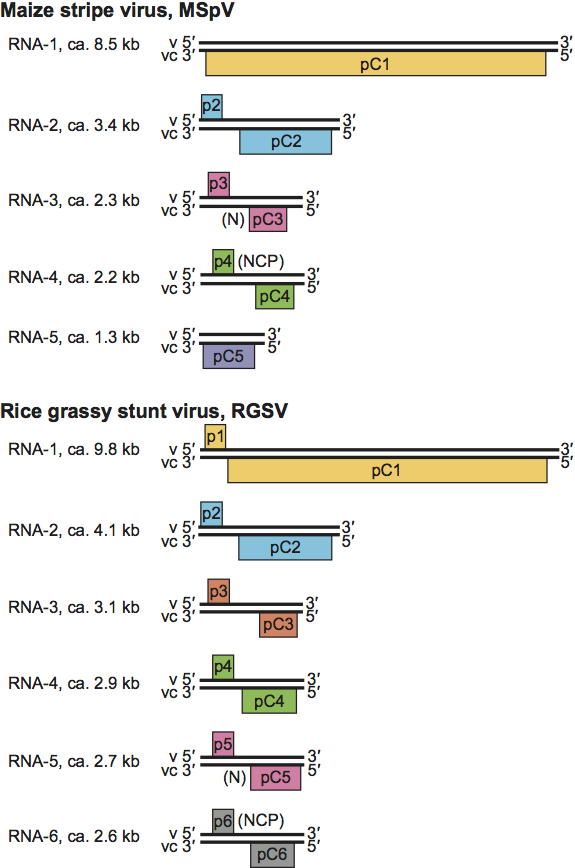
Figure 3 Diagram of the expression of the ambisense RNA of tenuiviruses. The black circle signifies the Cap, the broken line signifies non-viral nucleotides, V signifies virion-sense RNA and VC signifies virion-complementary sense RNA.
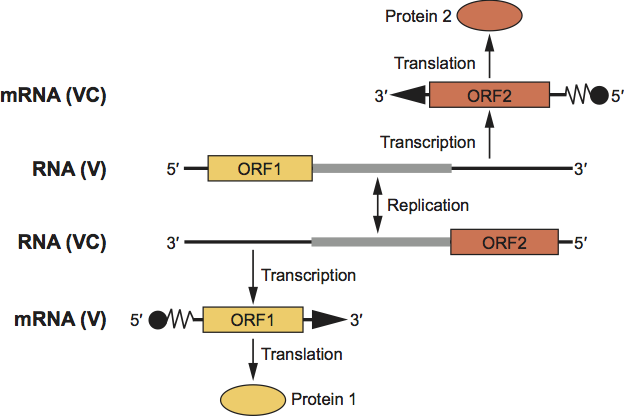
Figure 4 Phylogenetic tree showing the relationships among tenuiviruses. Input data were nt sequences of the coding regions of RNA-3 and -4 (RNA-5 and -6 respectively for RGSV) as aligned by PileUp (GCG-Wisconsin 9.0). A total of 3020 characters with clear positional homology across the alignment were used for the phylogenetic analysis. The phylogenetic tree was generated using Maximum Likelihood criteria as implemented by PAUP 4.0(b10), allowing for variable nucleotide substitution rates, rate heterogeneity between characters and rate heterogeneity between lineages. The tree was significantly superior to alternative trees, as determined by likelihood analysis. Bootstrap probabilities were calculated separately for the main tree and for the resolution among the strains of RHBV, RSV and RGSV. The following sequences were used: RHBV-co (AF004658, L14952); RHBV-cr (L07940, AF004657); EHBV (L75930, L48441); UHBV (U82447, U82446); RSV-jp(t) (X53563, D10979); RSV-jp(m) (D01094, D01039); RSV-cn (Y11095,Y11096); MSpV-us (M57426, L13438); RGSV-ph(l) (AB000403, AB000404); RGSV-ph(sc) (AB023779, AB023780); RGSV-cn (AF290947, AF287949).