Family: Orthomyxoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are spherical or pleomorphic, 80–120 nm in diameter. Newly isolated influenza viruses contain a significant proportion of filamentous forms, sometimes up to several micrometers in length, whereas laboratory strains with a long passage history in eggs or cell culture are represented mainly by spherical particles (Figure 1). The virion envelope is derived from the cell membrane, incorporating virus glycoproteins (one to three in number) and non-glycosylated proteins (one or two in number). Virion surface glycoprotein projections are 10–14 nm in length and 4–6 nm in diameter. The virus genome is segmented, has helical symmetry, and consists of different size ribonucleoproteins (RNP), 50–150 nm in length.
Physicochemical and physical properties
The virion molecular mass (Mr) is 250 106. Virion buoyant density in aqueous sucrose is 1.19 g cm−3. S20,w of non-filamentous particles is 700–800S. Virions are sensitive to heat, lipid solvents, non-ionic detergents, formaldehyde, irradiation and oxidizing agents.
Nucleic acid
Depending on the genus, virions contain different numbers of segments of linear, negative sense ssRNA: eight segments: influenza A virus (FLUAV), influenza B virus (FLUBV) and infectious salmon anemia virus (ISAV); seven segments: influenza C virus (FLUCV) and Dhori virus (DHOV); six segments: Thogoto virus (THOV). Segment lengths range from 736 to 2396 nt. Genome size ranges from 10.0 to 14.6 kb. RNA segments possess conserved and partially complementary 5′- and 3′-end sequences with promoter activity. Shorter viral RNA segments may occur in defective particles.
Proteins
Structural proteins common to all genera include: three polypeptides that form the viral RdRp (e.g., PA, PB1, PB2 in FLUAV); a nucleoprotein (NP), which is associated with each genome ssRNA segment to form the RNP; a hemagglutinin (HA, HE [HEF] or GP), which is an integral, type I membrane glycoprotein involved in virus attachment, envelope fusion and neutralization; and a non-glycosylated matrix protein (M1 or M). The HA of FLUAV is acylated at the membrane-spanning region and has N-linked glycans at a number of sites. In addition to its hemagglutinating and fusion properties, the HE (HEF) protein of FLUCV has esterase activity that functions as a receptor-destroying enzyme. In contrast, the GP of THOV is unrelated to influenzavirus proteins, but shows sequence homology to a baculovirus surface glycoprotein. Members of the genera Influenzavirus A and Influenzavirus B have an integral, type II envelope glycoprotein (neuraminidase, NA), which contains sialidase activity. Depending on the genus, viruses possess small integral membrane proteins (M2, NB, BM2, or CM2) that may be glycosylated. M2 and BM2 function as proton-selective ion channels in mammalian cells, acidifying the virion interior during uncoating and fusion and equilibrating the intralumenal pH of the trans-Golgi apparatus with that of the cytoplasm. The ion-channel activity of only the former is inhibited by the adamantane anti-influenza A drugs, amantadine and rimantadine. In addition to the structural proteins and depending on the genus, viruses may code for two nonstructural proteins (NS1, NS2 [NEP]) although NS2 is also found in the virus particle. Virion enzymes (variously represented and reported among genera) include a transcriptase (PB1 in influenzaviruses A, B, C and thogotoviruses), an endonuclease (PA in influenzaviruses A, B, C), and a receptor-destroying enzyme (neuraminidase (NA) for FLUAV and FLUBV, or 9-0-acetyl-neuraminyl esterase in the case of the FLUCV HE [HEF] protein).
Lipids
Lipids in the virion envelope constitute about 18–37% of the particle weight. They resemble lipids of the host cell plasma membrane.
Carbohydrates
Carbohydrates in the form of glycoproteins and glycolipids constitute about 5% of the particle weight. They are present as N-glycosidic side chains of glycoproteins, as glycolipids, and as mucopolysaccharides. Their composition is host- and virus-dependent.
Genome organization and replication
The genome codes for up to 12 proteins of 14–96 kDa. The five largest genome segments encode one protein each (with the exception of the PB1 segment, which for most (but not all) strains, encodes PB1 and PB1-F2 proteins and many also encode a polypeptide PB1 N40 translated in the same open reading frame as PB1 but initiating at a second AUG codon.). By contrast, smaller segments often code for additional proteins from spliced or bicistronic mRNAs. Generally the three largest RNAs encode the P proteins, and the 4th and 5th the viral HA (HE [HEF], GP) and NP proteins. Depending on the virus, the smaller RNA species encode the NA protein (FLUAV NA and FLUBV NA, NB: 6th RNA), the membrane proteins (FLUAV M1, M2 and FLUBV M1, BM2: 7th RNA; FLUCV M1, CM2 and THOV M, ML and DHOV M1: 6th RNA, ISAV: 8th RNA) and NS proteins (FLUAV and FLUBV NS1, NS2 [NEP]: 8th RNA; FLUCV NS1, NS2 [NEP]: 7th RNA; ISAV 7th RNA). Gene reassortment occurs during mixed infections involving viruses of the same genus, but not between viruses of different genera (e.g., FLUAV and FLUBV).
Virus entry involves the HA (HE [HEF], GP) and occurs by receptor-mediated endocytosis. The receptor determinant of influenzaviruses consists of sialic acid bound to glycoproteins or glycolipids. In endosomes, low pH-dependent fusion occurs between viral and cell membranes. For influenzaviruses, infectivity and fusion depend on the post-translational cleavage of the virion HA (FLUAV and FLUBV: HA1, HA2; FLUCV: HE1 HE2 [HEF1, HEF2]) protein to result in the production of a hydrophobic group of amino acids at the amino terminal of the HA2 molecule. Cleavability depends, among other factors, on the number of basic amino acids at the cleavage site. No requirement for glycoprotein cleavage has been demonstrated for the GP species of thogotoviruses. Integral membrane proteins migrate through the Golgi apparatus to localized regions of the plasma membrane. New virions form by budding, thereby incorporating matrix protein and the viral RNPs which align below regions of the plasma membrane containing viral envelope proteins. Budding is from the apical surface in polarized cells.
Viral RNPs are transported to the cell nucleus where the virion transcriptase complex synthesizes mRNA species. For influenzaviruses, mRNA synthesis is primed by capped RNA fragments 10–13 nt in length that are generated from host heterogeneous nuclear RNA species by viral endonuclease activity associated with the PB1 and PA proteins, after cap recognition by PB2. Thogotoviruses differ from influenzaviruses in having capped viral mRNA without host-derived sequences at the 5′ end. Virus-specific mRNA synthesis is inhibited by actinomycin D or α-amanitin due to inhibition of host DNA-dependent RNA transcription and a (presumed) lack of newly synthesized substrates that allow the viral endonuclease to generate the required capped primers. Virus-specific mRNA species are polyadenylated at the 3′ termini through a mechanism of iterative copying of an oligoU tract in the vRNA template. The mRNAs lack sequences complementary to the 5′-terminal (ca. 16) nucleotides of the viral RNA segment. Certain mRNAs are spliced to provide alternative products (Figure 2).
Protein synthesis occurs in the cytoplasm. However, NP, M1 and NS1 proteins accumulate in the cell nucleus during the first few hours of replication, then migrate to the cytoplasm. Cytoplasmic inclusions of NS1 may be formed.
Complementary RNA molecules which act as templates for new viral RNA synthesis are full-length transcripts and are neither capped nor polyadenylated. These RNAs exist as RNPs in infected cells.
Reverse genetics systems (technologies that allow one to genetically engineer viruses) have been established for FLUAV, FLUBV, FLUCV and THOV, allowing their generation entirely from cloned cDNA.
Antigenic properties
The best studied antigens are the NP, HA, NA, M1 and NS1 proteins of FLUAV and FLUBV. Considerable variation occurs among the FLUAV HA and NA antigens, less for FLUBV or the HE (HEF) surface antigens of FLUCV. THOV and DHOV do not cross-react in standard serologic tests, while DHOV and Batken virus do. Antibodies to HA, HE (HEF), or GP neutralize virus infectivity. Two major antigenic groups based on the properties of the HA have been identified in ISAV.
Influenza viruses agglutinate erythrocytes of many species. Serotype-specific antibodies may block agglutination. The NA or HE (HEF) of attached influenza virions may destroy sialic acid on the erythrocyte surface and the virus receptors, resulting in the elution of virus. Hemolysis of erythrocytes may be produced by HA at acid pH. In comparison to the influenzaviruses, thogotoviruses and isaviruses exhibit limited hemagglutination with certain erythrocyte species.
Biological properties
Certain influenzaviruses A naturally infect humans and cause respiratory disease. Particular influenzaviruses A infect other mammalian species and a variety of avian species. Interspecies transmission, though rare, is well documented. Influenza B virus strains appear to naturally infect mainly humans and cause epidemics every few years. Influenzaviruses C cause more limited outbreaks in humans and may also infect pigs. Human influenzaviruses A and B replicate in the amniotic cavity of embryonated hen eggs, and after adaptation they can also be propagated in the allantoic cavity. Influenzaviruses C replicate only in the amniotic cavity. Primary kidney cells from monkeys, humans, calves, pigs and chickens support replication of many FLUAV and FLUBV strains. The majority of these viruses require the addition of trypsin to the growth medium, so that proteolytic HA activation and multiple cycles of replication can occur in some continuous cell lines.
Natural transmission of influenzaviruses is by aerosol (human and most non-aquatic hosts) or is water-borne (waterfowl) but direct contact may also be important. Thogoto and Dhori viruses are transmitted by ticks and replicate in both ticks and a variety of tissues and organs in mammalian species as well as in mammalian cell cultures. In some laboratory species (e.g., hamsters for THOV) these infections have a fatal outcome. Unlike influenzaviruses, these viruses do not cause respiratory disease and do not replicate in embryonated hens’ eggs. Transmission of isaviruses is via water. Orthomyxoviruses have an Mx1-sensitive step in their multiplication cycle.
Genus Influenzavirus A
Type species Influenza A virus
Distinguishing features
Member viruses of the genus Influenzavirus A all have eight genome segments. The hemagglutinin and neuraminidase receptor-destroying enzyme are different glycoproteins. The conserved end sequences of the viral RNAs are 5′-AGUAGAAACAAGG and 3′-UCG(U/C)UUUCGUCC. The exact order of electrophoretic migration of the RNA segments varies with strain and electrophoretic conditions. On the basis of the gene sequences, for influenza A virus the segments 1–3 encoded PB2, PB1 and PA proteins are estimated to be 84 kDa (observed: 87 kDa), 87 kDa (observed: 96 kDa), and 83 kDa (observed: 85 kDa), respectively. RNA segment 2, the segment that encodes PB1 also encodes a second polypeptide read in an alternative reading frame, PB1-F2, which varies in length between viruses, the full length protein being in the order of 90 amino acids in length, some strains of virus encode a PB1-F2 of around 55 amino acids, the vast majority of the pandemic A(H1N1) 2009 viruses encode a truncated 11-amino acid PB1-F2. The same RNA segment of some viruses encodes a third polypeptide, PB1 N40, 718 amino acids in length. RNA segment 4 encoded (unglycosylated) HA is 63 kDa (glycosylated HA1 is 48 kDa, HA2 is 29 kDa). The segment 5 encoded NP is 56 kDa (observed: 50–60 kDa). The segment 6 encoded NA is 50 kDa (observed: 48–63 kDa). The segment 7 encoded M1 and M2 proteins are 28 kDa (observed: 25 kDa) and 11 kDa (observed: 15 kDa), respectively. The segment 8 encoded NS1 and NS2 (NEP) are 27 kDa (observed: 25 kDa) and 14 kDa (observed: 12 kDa), respectively.
Antigenic properties
Antigenic variation occurring within the HA and NA antigens of influenzaviruses A has been analyzed in detail. Based on antigenicity, 16 subtypes of HA and nine subtypes of NA are recognized for influenzaviruses A. Additional variation occurs within subtypes. By convention, new isolates are designated by their serotype/host species/site of origin/strain designation/year of origin and (HA [H] and NA [N] subtype); e.g. A/chicken/Novosibirsk/65/2005 (H5N1). In humans, continual evolution of new strains occurs, and older strains apparently disappear from circulation. The majority of neutralizing antibodies are directed to the HA. If NA antibody is present during multicycle replication, it inhibits virus release and reduces virus yield. Antibody to the amino terminus of M2 reduces virus yield in tissue culture.
Biological properties
Epidemics of respiratory disease in humans during the 20th–21st century have been caused by influenzaviruses A having the antigenic composition H1N1, H2N2 and H3N2. The pandemics of 1918, 1977 and 2009 were caused by H1N1 viruses, H2N2 caused “Asian influenza” in 1957 and in 1968 “Hong Kong influenza” was caused by an H3N2 virus. H1N2 reassortant viruses between H1N1 and H3N2 human viruses appeared in 2001 and became established, circulating viruses until 2004. Limited outbreaks of respiratory disease in humans caused by antigenically novel viruses occurred in 1976 in Fort Dix, New Jersey, when classical swine H1N1 viruses infected military recruits; and sporadic infections with swine H1N1 viruses have occurred in the intervening years. In 1997 and 2003 in Hong Kong H5N1 viruses caused outbreaks in poultry and contemporary illnesses and deaths in humans. The continued circulation of H5N1 viruses in birds has been associated with zoonotic infections of humans with H5N1 viruses, with a large proportion proving fatal. H9N2 viruses present in poultry have caused occasional illness in humans in China, first observed in 1998, and zoonotic infections have continued to be documented. Influenzaviruses A of subtype H7N7 and H3N8 (previously designated equine 1 and equine 2 viruses, respectively) cause outbreaks of respiratory disease in horses; but H7N7 virus has not been isolated from horses since the late 1970s. Influenzaviruses A (H1N1) and (H3N2) have been isolated frequently from pigs. The H1N1 viruses isolated from swine in recent years appear to be of three general categories: those closely related to classical “swine influenza” and which cause occasional human cases; those first characterized in samples collected from swine in 1979 and genetically more closely related in all gene segments to H1N1 viruses isolated in birds which have become established and cause infection among pigs in Europe and Asia; and those resembling viruses isolated from epidemics in humans since 1977. Swine H1N1 viruses have also been observed as triple reassortants with genes originating from the swine pool, the avian pool and the human pool; the “triple” reassortants with genes form three distinct gene pools were first observed in swine H3N2 viruses. H3N2 viruses from swine appear to contain HA and NA genes closely related to those from human epidemic strains. Triple gene reassortant viruses possessing the H3 HA and N2 NA from a recent human virus and other genes from a swine and/or avian virus were first identified in the North American pig population in 1998 and have been circulating since then. Infections of pigs with the A(H1N1) 2009 pandemic have been documented associated with reverse zoonosis; H1N2 viruses, distinct from those in humans, have been isolated from pigs in UK, France, Japan and the US. Influenzaviruses A (H7N7 and H4N5) have caused outbreaks in seals, with virus spread to non-respiratory tissues in this host. H7N7 viruses have been isolated from conjunctival infections of a laboratory worker and a farm worker in 1980 and 1996, respectively and from humans involved in disease control in 2003. In addition, in 2003, a human was fatally infected with a highly pathogenic avian influenza H7N7 virus. Pacific Ocean whales have reportedly been infected with type A (H1N3) virus. Other influenza subtypes have also been isolated from lungs of Atlantic Ocean whales off North America. FLUAV (H10N4 and H3N2) has caused outbreaks in mink. All subtypes of HA and NA, in many different combinations, have been identified in isolates from avian species, particularly wild aquatic birds, chickens, turkeys and ducks. Pathology in avian species varies from non-apparent infection (often involving replication in, and probable transmission via, the intestinal tract), to more severe infections (observed with subtypes H5 and H7) with spread to many tissues and high mortality rates. The structure of the HA glycoprotein, in particular the specificity of its receptor binding site and its cleavability by host protease(s), appears to be critical in determining the host range and organ tropisms of influenza viruses. The NS1 also contributes to the outcome of infection by mitigating host defense mechanisms; e.g., through anti-interferon activity. In addition, interactions between gene products determine the outcome of infection. Interspecies transmission apparently occurs in some instances without genetic reassortment (e.g., the direct transmission of H1N1 virus from swine to humans and vice versa, H3N2 virus from humans to swine, and the recent transmissions of H5N1 and H9N2 viruses from poultry to humans). In other cases, interspecies transmission may involve RNA segment reassortment in hosts infected with more than one strain of virus, each with distinct host ranges, or epidemic properties (e.g., 1968 isolates of H3N2 viruses were derived by reassortment between a human H2N2 virus and a virus containing an H3 HA). Laboratory animals that may be infected with influenzaviruses A include ferrets, mice, hamsters and guinea pigs, as well as some small primates such as squirrel monkeys.
Species demarcation criteria in the genus
Only a single species is currently recognized in the genus Influenzavirus A. The species is comprised of a cluster of strains that replicate as a continuous lineage and can genetically reassort with each other. Therefore, although 16 different HA subtypes and nine different NA subtypes are recognized among influenzaviruses A replicating in birds, separate species designations have not been accorded to these subtypes. All isolates are capable of exchanging of RNA segments (reassortment).
List of species in the genus Influenzavirus A
| Influenza A virus |
|
|
| Influenza A virus A/PR/8/34 (H1N1) | [J02144, J02146, J02148, J02151, V00603, V01099, V01104, V01106] | (FLUAV- A/PR/8/34 (H1N1)) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Influenzavirus A but have not been approved as species
None reported
Genus Influenzavirus B
Type species Influenza B virus
Distinguishing features
Member viruses of the genus Influenzavirus B all have eight genome segments. As for members of the genus Influenzavirus A, hemagglutinin and the neuraminidase receptor-destroying enzyme are different glycoproteins. The conserved end sequences of the viral RNAs of the influenzaviruses B are 5′-AGUAG(A/U)AACAA and 3′-UCGUCUUCGC. Influenza B virus proteins have sizes similar to those for influenza A virus. NB: the second product of FLUBV segment 6 is 11 kDa (glycosylated 18 kDa).
Antigenic properties
Antigenic variation within the HA and NA antigens of influenzaviruses B has also been analyzed in detail. In contrast to influenzaviruses A, no distinct antigenic subtypes are recognized for members of the species Influenza B virus, however, viruses with antigenically and genetically distinguishable lineages of HA and NA (e.g., the B/Victoria/2/87-like and the B/Yamagata/16/88-like viruses) have co-circulated in humans for over two decades. Influenzaviruses B infect humans and they are designated by their serotype/site of origin/strain designation/year of origin (as above B/Victoria/2/87 and B/Yamagata/16/88). There is a report of influenza B infection of marine mammals (seals) and a single virus isolate has been reported; like for influenza A viruses, the species of origin is included for virus strains collected from animals (e.g. B/Seal/Netherlands/1/99). Most neutralizing antibodies bind the HA protein.
Biological properties
Influenzaviruses B, first isolated in 1940, have been circulating continuously in humans and causing recurrent epidemics of respiratory disease. Antigenic change (antigenic drift) occurs more slowly among influenzaviruses B than influenzaviruses A.
Species demarcation criteria in the genus
Only a single species is currently recognized in the genus Influenzavirus B. The species is comprised of a cluster of strains that replicate as a continuous lineage and can reassort genetically with each other. Although considerable antigenic and sequence differences exist among viruses in this genus, these differences are not sufficient for designation of separate species.
List of species in the genus Influenzavirus B
| Influenza B virus |
|
|
| Influenza B virus B/Lee/40 | [J02094, J02095, J02096, K00423, K01395, M20168, M20170, M20172] | (FLUBV- B/Lee/40)) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Influenzavirus B but have not been approved as species
None reported.
Genus Influenzavirus C
Type species Influenza C virus
Distinguishing features
Member viruses of the genus Influenzavirus C naturally infect humans. Viruses have seven genome segments. They lack neuraminidase. The HE (HEF) protein contains the receptor binding and fusion activities and also functions as the receptor-destroying enzyme, 9-0-acetylneuraminyl esterase. The conserved end sequences of the viral RNAs of the influenzaviruses C are 5′-AGCAG(U/G)AGCAAG and 3′-UCGUCUUCGUC. RNA segments 1–3 encode the P proteins (87.8 kDa, 86.0 kDa, and 81.9 kDa, respectively). Segment 4 encodes HE (HEF) (unglycosylated: 72.1 kDa, glycosylated: 88.0 kDa), segment 5 NP (63.5 kDa), segment 6 M1 and P42 (27.0 kDa and 42.0 kDa, respectively) and segment 7 NS1 (27.7 kDa) and NS2 (NEP) (21.0 kDa). Proteolytic cleavage of P42 at an internal signal peptidase cleavage site gives rise to M1’ (p31) and CM2 proteins (31.0 kDa and 18.0 kDa, respectively).
Antigenic properties
Antigenic drift characterized by the emergence of successive antigenic variants which have descended from those that circulated previously apparently does not occur among influenzaviruses C; however, antigenic variation between distinct co-circulating lineages has been detected in HI tests with both anti-HE (HEF) Mabs and polyclonal antisera. Viruses exhibit no cross-reactivity with influenzaviruses A and B, although homologies of HE (HEF) to influenzavirus A and B HA were identified near the amino and carboxy termini and several of the cysteines co-aligned in the sequences. Antibody to HE (HEF) neutralizes infectivity. Influenzaviruses C are designated by their serotype/site of origin/strain designation/year of origin, e.g. C/Catalonia/1457/2009, and the host from which the viruses was isolated when not human, e.g. C/swine/Beijing/32/81.
Biological properties
Infection in humans is common in childhood. Occasional outbreaks, but not epidemics, have been detected. Swine in China have been reported to be infected by viruses similar to human Influenza C virus strains.
Species demarcation criteria in the genus
Only a single species is currently recognized in the genus Influenzavirus C. The species is comprised of a cluster of strains that replicate as a continuous lineage and can reassort genetically with each other. Although detectable antigenic and sequence differences exist among this genus, these differences are not sufficient for separate species designation.
List of species in the genus Influenzavirus C
| Influenza C virus |
|
|
| Influenza C virus C/Ann Arbor/1/50 | [AB126191, AB126192,AB126193, AB126194,AB126195, AB126196, AB283001] | (FLUCV- C/Ann Arbor/1/50) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Influenzavirus C but have not been approved as species
None reported.
Genus Thogotovirus
Type species Thogoto virus
Distinguishing features
Morphology and morphogenesis of these viruses show similarities with the influenzaviruses. Virions contain segments of linear, negative sense ssRNA. Total genomic size is about 10 kb. Sequences of the ends of vRNA are partially complementary and resemble those of influenzaviruses. The conserved end sequences of THOV viral RNAs are 5′-AGAGA(U/A)AUCAA(G/A)GC and 3′-UCGUUUUUGU(C/U)CG (segments 1-5) or 3′-UCACCUUUGUCCG (segment 6). Intrastrand base-pairings are favored over interstrand base-pairings, leading to a “hook-like” or cork-screw structure. THOV RNA segments 1-3 encode PB2, PB1 and PA proteins (88, 81 and 71.5 kDa, respectively) that exhibit homology to the respective influenzavirus proteins. The single glycoprotein GP (THOV: 75 kDa; DHOV: 65 kDa) is encoded by the fourth segment. It is unrelated to any influenzavirus protein but shows amino acid sequence similarity with the glycoprotein gp64 of baculoviruses. The fifth segment encodes the NP (THOV: 52 kDa; DHOV: 54 kDa), which is related to influenzavirus NP. The sixth segment of THOV encodes the matrix protein M (29 kDa, translated from a spliced mRNA) and a second protein ML (32 kDa, translated from the unspliced mRNA). ML represents an elongated version of M with a C-terminal extension of 38 aa. The sixth segment of DHOV encodes the M1 protein (30 kDa) and may encode another protein, M2 (15 kDa, but not detected in virions) of unknown function. The coding of a putative seventh segment of DHOV is not known.
Antigenic properties
Antigenic relationships between THOV and DHOV viruses are not apparent and none of the virus proteins are related antigenically to those of influenzaviruses; however, serological cross-reactivity between DHOV and Batken virus has been demonstrated. For THOV and DHOV, several viruses have been isolated; however, the relationships of these isolates to the prototype viruses are not known.
Biological properties
THOV and DHOV are transmitted between vertebrates by ticks. Comparatively low levels of hemagglutination occur at acidic pH and not at physiological pH. No receptor-destroying enzyme has been observed. Fusion of infected cells occurs at acidic pH indicating that cell entry is via the endocytic pathway as for the influenzaviruses. Fusion is inhibited by neutralizing monoclonal antibodies directed against GP. Replication is inhibited by actinomycin-D or α-amanitin. Nucleoprotein accumulates early in replication within the nucleus. M of THOV is required for the generation of virus-like particles and infectious recombinant viruses by reverse genetics. In contrast, ML is dispensable for virus growth in cell culture, but appears to be a virulence factor with interferon antagonistic function. THOV is inhibited by the interferon-induced Mx GTPases at an early step in the virus multiplication cycle. Reassortment between THOV temperature-sensitive mutants has been demonstrated experimentally in co-infected ticks and in vertebrates.
Species demarcation criteria in the genus
THOV has been isolated from Boophilis sp. and Rhipicephalus sp. ticks in Kenya and Sicily, from Amblyomma variegatum ticks in Nigeria, and from Hyalomma sp. ticks in Nigeria and Egypt. THOV is known to infect humans in natural settings, and serological evidence suggests that other animals (including cattle, sheep, donkeys, camels, buffaloes and rats) are also susceptible to this virus. THOV has been isolated in the Central African Republic, Cameroon, Uganda, and Ethiopia as well as in southern Europe. DHOV has a somewhat different, but overlapping geographic distribution that includes India, eastern Russia, Egypt and southern Portugal. DHOV has been isolated from Hyalomma sp. ticks. As demonstrated by the accidental infection of laboratory workers, DHOV is able to infect humans, causing a febrile illness and encephalitis. Serologic evidence suggests that cattle, goats, camel and waterfowl are also susceptible to this virus. There is no detectable serological reactivity between THOV and DHOV and the sequence diversity of 37% and 31% in the nucleoprotein and the envelope protein, respectively, argues for separate species status. Batken virus isolated from mosquitoes and ticks from Russia cross-reacts serologically with DHOV and shares 98% identity in a portion of the nucleoprotein and 90% identity in a portion of the envelope protein. These data suggest that Batken virus, although isolated from both mosquitoes and ticks, is closely related to DHOV.
List of species in the genus Thogotovirus
| Thogoto virus |
|
|
| Thogoto virus strain SiAr 126 | [NC_006508, NC_006495, NC_006496, NC_006506, NC_006507, NC_006504] | (THOV) |
| Dhori virus |
|
|
| Dhori virus isolate Dhori/1313/61 | [GU969308, GU969313, GU969309, GU969310, GU969311, GU969312] | (DHOV) |
| Batken virus strain LEIV306K | [X97340, X97341] | (BATV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Thogotovirus but have not been approved as species
Araguari virus (ARAV)
Genus Isavirus
Type species Infectious salmon anemia virus
Distinguishing features
Isaviruses are similar in morphology to influenzaviruses. Surface projections are 10 nm in length. Virion surface glycoproteins are the hemagglutinating and receptor-destroying glycoprotein (HE) and the fusion glycoprotein (F). The receptor-destroying function is an acetylesterase. Isaviruses have eight linear genome segments of negative sense ssRNA. The vRNA 5′-AGUAAAAA(A/U) and 3′-UCG(U/A)UUCUA terminal sequences are conserved among isaviruses and partially complementary, with some sequence resemblance to those of influenzaviruses. Total genome size is about13.5 kb. Each of the two smallest segments encodes two proteins. Segment 7 gives a spliced mRNA product. The synthesis of mRNA is primed by RNA fragments 8–18 nt in length. The genes of segments 1, 2 and 4 are thought to encode the P proteins based on limited homologies to other RdRp; estimated sizes are 79.9, 80.5, and 65.3 kDa, respectively. Segment 3 encodes NP, which is phosphorylated (68 kDa). Segment 5-encodes F (48.8 kDa), a type 1 membrane glycoprotein. F is proteolytically processed into two disulfide-linked subunits F1 and F2. Segment 6 encodes HE, a type 1 membrane glycoprotein (42.7 kDa). Segment 7 encodes a non-structural protein (NS) with interferon antagonistic function and a protein of unknown function, with estimated sizes of 34.2 and 17.6 kDa, respectively. Segment 8 encodes the matrix protein (M1) and a RNA binding protein with estimated sizes of 22 and 27.6 kDa, respectively.
Antigenic properties
There is no known antigenic relationship between isavirus proteins and those of influenzaviruses. Neutralizing antibodies are mainly directed against the HE (HEF) protein. The humoral immune response of the host recognizes mainly the HE and NP. There are many isolates of ISAV, and they have been divided into two major antigenic groups based on properties of the HE.
Biological properties
ISAV is transmitted through water. It agglutinates erythrocytes of many fish species, but not avian or mammalian erythrocytes. Fusion of the virus with infected cells occurs at low pH, suggesting endocytic cell entry. The maximum rate of virus replication in the salmon head kidney cell line (SHK-1) is in the temperature range 10–15 °C; at 20 °C the production of infectious virus is reduced by more than 99% and no replication is observed at 25 °C. Replication is inhibited by actinomycin D and α-amanitin.
Species demarcation criteria in the genus
Only a single species is currently recognized in the genus Isavirus. The species is comprised of a cluster of strains that replicate as a continuous lineage and can reassort genetically with each other. Although detectable antigenic and sequence differences exist among this genus, these differences are not sufficient for separate species designation.
List of species in the genus Isavirus
| Infectious salmon anemia virus |
|
|
| Infectious salmon anemia virus | [AF404341, AF404340, AF404346, AF404345, AF404344, AF404343, AY373381, AF404342] | (ISAV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Isavirus but have not been approved as species
None reported.
List of other related viruses which may be members of the family Orthomyxoviridae but have not been approved as species
| Quaranfil virus | [FJ861695, FJ861694, GQ499304, GQ499303, GQ499302] | (QRFV) |
| Johnston Atoll Virus | [FJ861697, FJ861696] |
|
| Lake Chad Virus | [FJ861698] |
|
It has been suggested that Quaranfil virus might represent a novel genus in the family Orthomyxoviridae. The virus was originally isolated from two children with febrile illness from the villages of Quaranfil and Sindbis in Egypt in 1953. Several strains of Quaranfil virus have been isolated from ticks and seabirds in multiple countries throughout Africa and the Middle East. Johnston Atoll virus is serologically related to Quaranfil. It was originally isolated from ticks (Ornithodoros capensis) collected in 1964 from a Noddy Tern (Anous stolidus) nest, Sand Island, Johnston Atoll in the central Pacific. Morphology and morphogenesis of these viruses show similarities with the influenzaviruses. Quaranfil virions are reported to contain at least six segments of linear, negative sense ssRNA which have been completely sequenced. Sequences of the ends of vRNA are partially complementary and resemble those of influenzaviruses. The conserved end sequences of both Quaranfil and Johnston Atoll viral RNAs are 5′-AGCAAUCACAA and 3′-UCGUUAGUGU(A/U)(A/G). Quaranfil RNA segments 1–3 (2421 nt, 2404 nt and 2386 nt) encode single ORFs exhibit protein domain homology to the respective influenzavirus polymerase proteins PB2, PA and PB1. The fifth segment (1616 nt) is unrelated to any influenzavirus protein but shows aa sequence similarity with the glycoprotein (gp64) of baculoviruses and the GP of the thogotoviruses. The fourth segment (1726 nt) contains one single predicted ORF of 527 aa which does not share significant sequence homology with any protein currently in the GenBank database. Similarly, the sixth segment of 898 nt contains one predicted ORF of 266 aa which does not share sequence homology with any known protein. It is unclear which segments encode the nucleoprotein or matrix protein of the Quaranfil viruses. There is no significant antigenic relationship between Quaranfil and either the thogotoviruses or the influenzaviruses. Quaranfil and Johnston Atoll are transmitted between vertebrates by ticks. Comparatively low levels of hemagglutination occur at acidic pH and not at physiological pH for Quaranfil viruses against goose red blood cells. Johnston Atoll virus does not agglutinate goose red blood cells across pH 5.75–7.0.
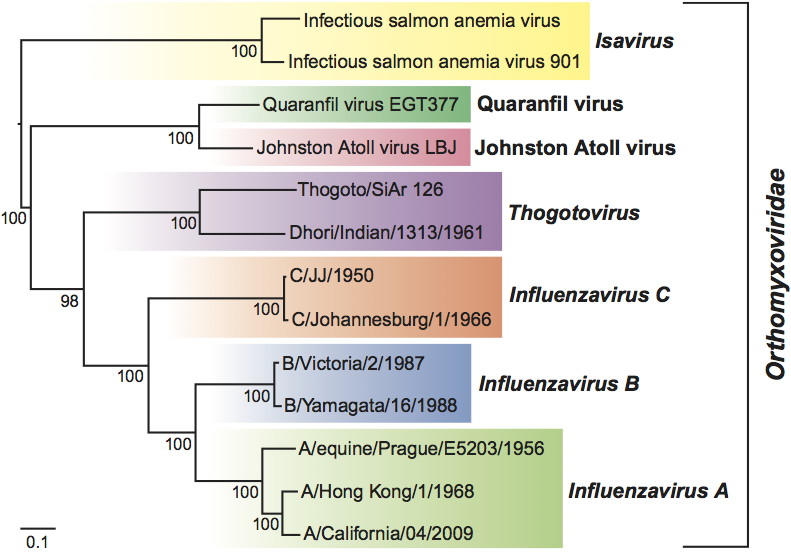
Phylogenetic relationships within the family
Phylogenetic relationships within the family are illustrated in Figure 3.
Similarity with other taxa
Not reported.
Derivation of names
Influenza: Italian form of Latin influentia, “epidemic”, originally used because epidemics were thought to be due to astrological or other occult “influences”.
Isavirus: from infection salmon anemia virus
Myxo: from Greek myxa, “mucus”.
Ortho: from Greek orthos, “straight”.
Thogoto: from Thogoto Forest near Nairobi, Kenya, where Thogoto virus was first isolated from ticks.
Further reading
Journals and books
Clerx et al., 1983 J. Clerx, F. Fuller, D. Bishop, Tick-borne viruses structurally similar to orthomyxoviruses. Virology. 127 (1983) 205–219.
Falk et al., 1997 K. Falk, E. Namork, E. Rimstad, S. Mjaaland, B.H. Dannevig, Characterization of infectious salmon anemia virus, an orthomyxo-like virus isolated from Atlantic salmon (Salmo salar L.). J. Virol. 71 (1997) 9016–9023.
Kawaoka, 2006 Y. Kawaoka, Influenza Virology: Current Topics. In: Y. Kawaoka, Influenza Virology: Current Topics. Caister Academic Press, Wymondham, England2006.
Klenk et al., 2008 H.-D. Klenk, M.N. Matrosovich, J. Stech, Avian Influenza. Monographs in Virology 27. In: H.-D. Klenk, M.N. Matrosovich, J. Stech, Avian Influenza. Monographs in Virology 27. Karger, Basle2008.
Lamb and Horvath, 1991 R.A. Lamb, C.M. Horvath, Diversity of coding strategies in influenza viruses. Trends Genet. 7 (1991) 261–266.
Mettenleiter, 2009 Mettenleiter, T.C. (Ed.) (2009). Avian influenza. Rev. Sci. Tech. Off. Int. Epiz., 28(1), 203-217.
Mjaaland et al., 1997 S. Mjaaland, E. Rimstad, K. Falk, B.H. Dannevig, Genomic characterization of the virus causing infectious salmon anemia in Atlantic salmon (Salmo salar L.): an orthomyxo-like virus in a teleost. J. Virol. 71 (1997) 7681–7686.
Palese and Shaw, 2007 P. Palese, M.L. Shaw, D.M. Knipe, P.M. Howley, Orthomyxoviridae: The viruses and their replicationFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott Williams & Wilkins, Philadelphia20071647–1689.
Presti et al., 2009 R.M. Presti, G. Zhao, W.L. Beatty, K.A. Mihindukulasuriya, A.P. da Rosa, V.L. Popov, R.B. Tesh, H.W. Virgin, D. Wang, Quaranfil, Johnston Atoll, and Lake Chad viruses are novel members of the family Orthomyxoviridae. J. Virol. (83) (2009) 11599–11606.
Rimstad et al., 2001 E. Rimstad, S. Mjaaland, M. Snow, A.B. Mikalsen, C.O. Cunningham, Characterisation of the genomic segment of infectious salmon anemia virus that encodes the putative hemagglutinin. J. Virol. 75 (2001) 5352–5356.
Wise et al., 2009 H.M. Wise, A. Foeglein, J. Sun, R.M. Dalton, S. Patel, W. Howard, E.C. Anderson, W.S. Barclay, P. Digard, A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J. Virol. 83 (2009) 8021–8031.
Websites
NCBI Influenza Virus Resource: http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html
Influenza Research Database: http://www.fludb.org/brc/home.do?decorator=influenza
Global Initiative on Sharing All Influenza Data (GISAID): http://platform.gisaid.org/
Contributed by
McCauley J.W., Hongo S., Kaverin N.V., Kochs G., Lamb R.A., Matrosovich M.N., Perez D.R., Palese P., Presti R.M., Rimstad E. and Smith, G.J.D.
Figures
Figure 1 (Top) Diagram of an influenza A virus (FLUA) virion in section. The indicated glycoproteins embedded in the lipid membrane are the trimeric hemagglutinin (HA), which predominates, and the tetrameric neuraminidase (NA). The envelope also contains a small number of M2 ion channel proteins. The internal components are the M1 (matrix) protein and the viral ribonucleoprotein (RNP) consisting of RNA segments, associated nucleocapsid protein (NP), and the PA, PB1 and PB2 polymerase proteins. NS2 (NEP), also a virion protein, is not shown. (Bottom) Frozen-hydrated images of a spherical A/Aichi/2/68 X-31 virion (left) and a filamentous A/Udorn/72 virion (right)
(image courtesy of Peter Rosenthal, NIMR, London).
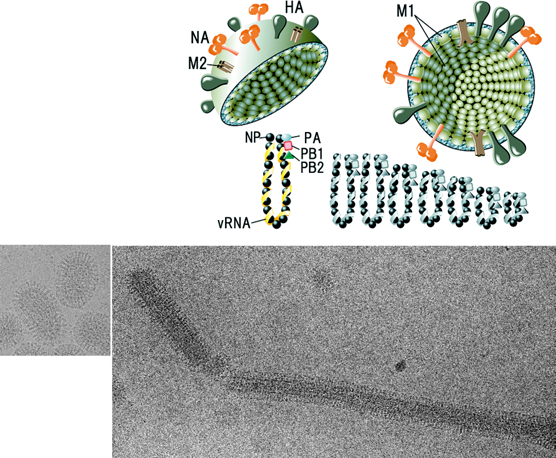
Figure 2 Orthomyxovirus genome organization. The genomic organization and ORFs are shown for genes that encode multiple proteins. Segments encoding the polymerase, hemagglutinin and nucleoprotein genes are not depicted as each encodes a single protein. (A) Influenza A virus PB1 segment ORFs. Initiation of PB1 translation is thought to be relatively inefficient based on Kozaks rule, likely allowing initiation of PB1-F2 translation by ribosomal scanning and results in PB1-F2 proteins of different size. In addition, the use of a second AUG, present in many but not all viruses, in frame in the PB1 ORF as the initiation codon encodes the polypeptide PB1 N40, the C terminal 718 amino acids of PB1. (B) Influenza A virus segment 7 showing M1 and M2 mRNAs and their coding regions. M1 and M2 share 9 amino-terminal residues, including the initiating methionine; however, the ORF of M2 mRNA (nt 7401004) differs from that of M1. (C) Influenza A virus segment 8 showing NS1 and NS2 (NEP) mRNAs and their coding regions. NS1 and NS2 (NEP) share 10 amino-terminal residues, including the initiating methionine. The ORF of NS2 (NEP) mRNA (nt 529-861) differs from that of NS1. (D) ORFs in Influenza B virus RNA segment 6, illustrating the overlapping reading frames of NB and NA. Nucleotide sequence surrounding the 2 AUG initiation codons, in the mRNA sense, is shown. (E) Influenza B virus RNA segment 7 ORFs and the organization of the ORFs used to translate the M1 and BM2 proteins. A stopstart pentanucleotide, thought to couple translation between the two ORFs, is illustrated. (F) Influenza C virus mRNAs derived from RNA segment 6. The unspliced and spliced mRNAs encode P42 and M1, respectively. The cleavage of P42 by a signal peptidase produces M1(p31) and CM2. (G) Thogoto virus segment 6 showing M and ML. M is translated from a spliced mRNA with a stop codon that is generated by the splicing process itself, as in Influenza C virus M1 mRNA. ML is translated from the unspliced transcript and represents an elongated form of M with a C-terminal extension of 38 aa. (H) Isavirus mRNAs derived from segment 7. The unspliced mRNA encodes the NS protein and the spliced mRNA encodes a polypeptide of unknown function. (I) Isavirus mRNAs derived from segment 8. ORF1 starts at nucleotide 22 and encodes the M1 protein, ORF2 starts at nucleotide 36, in the+2 reading frame relative to ORF1 and encodes a polypeptide of unknown function. For all panels, the boxes represent different coding regions. Introns in the mRNAs are shown by the V-shaped lines; filled rectangles at the 5 ends of mRNAs represent heterogeneous nucleotides derived from cellular RNAs that are covalently linked to viral sequences. Lines at the 5 and 3 termini of the mRNAs represent untranslated regions.
(Modified from Lamb and Horvath (1991). Trends Genet., 7, 261-266 and Garcia-Rosado et al. (2008). Virus Res., 133, 228-238.)

Figure 3 Phylogenetic relationships within the family Orthomyxoviridae. Nucleotide sequences of the polymerase basic 1 proteins (PB1) were aligned using transAlign and CLUSTAL W, and their phylogenetic relationships were determined by the neighbor-joining method (HKY model) using PAUP* (version 4.0b). The tree was mid-point rooted and bootstrap values (1000 replicates) are indicated on the branches. The GenBank accession numbers for the sequences used for comparison were (top to bottom) AF404346, GU830904, FJ861695, FJ861697, AF004985, M65866, M28060, AF170575, CY018763, CY018771, GU053121, CY044267 and FJ966080.