Family: Ophioviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Distinguishing features
Single stranded negative/possibly ambisense RNA genome (11.3–12.5 kb size) divided into 3–4 segments, each encapsidated in a single coat protein (43–50 kDa) forming filamentous virions about 3 nm in diameter, in shape of kinked (probably internally coiled) circles of at least two different contour lengths.
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Ophiovirus
Type species Citrus psorosis virus
Virion properties
Morphology
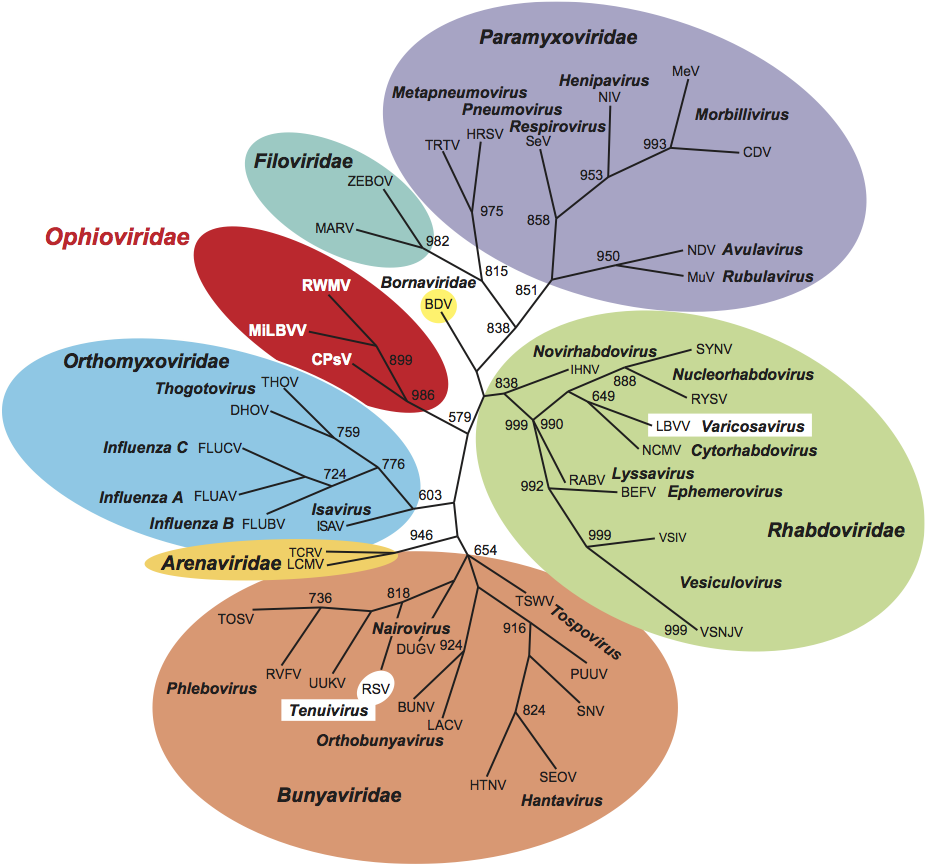
The virions are naked filamentous nucleocapsids about 3 nm in diameter (Figure 1), forming kinked (probably internally coiled) circles of at least two different contour lengths, the shortest length about 760 nm. The circles can collapse to form pseudo-linear duplex structures about 9–10 nm in diameter.
Physicochemical and physical properties
The Mr and sedimentation coefficients of virions are not known. The particles are unstable in CsCl but the buoyant density in cesium sulfate (for both Ranunculus white mottle virus [RWMV], and Mirafiori lettuce big-vein virus [MiLBVV]) is 1.22 g cm−3. The particles have limited stability between pH 6 and 8. Infectivity does not survive in crude sap held at 50 °C for 10 min (40–45 °C for tulip mild mottle mosaic virus, TMMMV). Particle structure survives limited treatment with organic solvents and nonionic or zwitterionic detergents.
Nucleic acid
The ssRNA genome is 11.3–12.5 kb in size and consists of three or four segments. Viral RNA of both polarities is present in purified virion preparations. As virions appear circularized, the presence of panhandle structures is suggested; however, significant complementation between the 5′- and the 3′-terminal sequences of genomic RNAs has not been demonstrated for either citrus psorosis virus (CPsV) or MiLBVV. At the 5′ and 3′ ends of MiLBVV genomic RNAs there are orthomyxovirus-like palindromic sequences that could fold into a symmetrically hooked conformation. These structures could not be predicted for CPsV. The size of RNA-1 is 8.2 kb for CPsV, about 7.5 kb for RWMV, 7.8 kb for MiLBVV and 7.6 kb for lettuce ring necrosis virus (LRNV). RNA-2 is about 1.8 kb for RWMV, MiLBVV and LNRV, 1.6 kb for CPsV and 1.7 for Freesia sneak virus (FreSV). RNA-3 is about 1.5 kb. For MiLBVV and LRNV a fourth genomic RNA of about 1.4 kb has been reported.
Proteins
There is one CP, varying in size according to species; 48–50 kDa for CPsV, about 43 kDa for RWMV, about 47 kDa for TMMMV, 48.5 kDa for MiLBVV, 48.4 kDa for FreSV and 48 kDa for LRNV.
Lipids
Not reported.
Carbohydrates
Not reported.
Genome organization and replication
All ophiovirus genome segments appear to be of negative polarity (Figure 2). RNA-1 contains a large ORF coding for a protein that contains the core polymerase module with the five conserved motifs proposed to be part of the RNA-dependent RNA polymerase (RdRp) active site. A paramyxovirus-RdRp domain (pfam 00946) is also present and the SDD sequence, a signature for segmented negative-stranded RNA viruses (Orthomyxoviridae, Arenaviridae and Bunyaviridae), occurs in motif C. A further small ORF has also been reported. RNA-2 contains a single ORF on the complementary strand for CPsV, LRNV and FreSV; for MiLBVV two ORFs are reported: the small second ORF present in the virion sense strand has not yet been confirmed as functional. RNA-3 codes for the CP. RWMV CP antigen accumulates in the cytoplasm of parenchyma cells. RNA-4 (ca. 1.4 kb) has been identified only from MiLBVV and LRNV. While RNA-4 of LRNV potentially encodes only one protein (ca. 38 kDa) of unknown function, that of MiLBVV contains two ORFs (37 and 10.6 kDa) overlapping by 38 nt. The second ORF lacks an initiation codon and is proposed to be expressed by a +1 translational frameshift. Nuclear localization signals (NLS) are reported for the CPsV, MiLBVV and RWMV polymerases and for the RNA-2-encoded proteins of CPsV and MiLBVV.For CPsV the 5′ ends of the complementary strand of RNA-1, RNA-2 and RNA-3 are identical, GATAC(T)7, but the 3′ ends are all different. For MiLBVV, the respective 5′ and 3′ ends are conserved among the four viral RNA segments.
Antigenic properties
The CP is the only significant antigenic element. In Western blots, CPs of RWMV, TMMMV, MiLBVV and LRNV appear to be slightly to moderately serologically related. CPsV CP is serologically unrelated to the others.
Biological properties
Ophioviruses can be mechanically transmitted to a limited range of test plants, inducing local lesions and systemic mottle. The natural hosts of CPsV, RWMV, MiLBVV and LRNV are dicotyledonous plants of widely differing taxonomy. In contrast, monocots are naturally infected by TMMMV [tulip (Tulipa gesneriana L., Liliaceae)] and FreSV [freesia (Freesia refracta hybrids, Iridaceae) and Lachenalia (Lachenalia hybrids, Hyacinthaceae)]. CPsV is commonly transmitted by vegetative propagation of the host and no natural vectors have been identified, although natural dispersion of psorosis disease has been observed in a few locations. No vector is known for RWMV. The zoospores of Olpidium brassicae transmit TMMMV, MiLBVV and LRNV; FreSV is soil-transmitted, presumably by the same vector.
CPsV has a wide geographical distribution in citrus in the Americas, in the Mediterranean and in New Zealand. RWMV has been reported in two species of the family Ranunculacae from Northern Italy, and in lettuce in France and Germany. TMMMV has been reported in tulips in Japan. MiLBVV is the causal agent of big-vein symptoms (zones cleared of chlorophyll, parallel to the veins) in lettuce; this widespread and damaging disease probably occurs world-wide. LRNV is closely associated with lettuce ring necrosis disease in The Netherlands, Belgium and France. FreSV has been reported in Europe, Africa, North America and New Zealand.
Species demarcation criteria in the genus
The different criteria considered for species demarcation in the genus are:
- differences in CP size
- no or distant serological relationship between CPs
- differences in natural host range
- different number, organization and/or size of genome segments.
Alignments between CPsV, FreSV, MiLBVV, LRNV and RWMV (partial cds) CP amino acid sequences show 31–52% identity (53–70% similarity) for isolates belonging to different species, while showing about 92–100% identity for isolates of the same species. Since CPs of MiLBVV and TMMMV (partial cds) share about 80% amino acid sequence identity, this may warrant the establishment of the following molecular criterion for ophiovirus species demarcation:
CP amino acid sequence identity of <85%.
List of species in the genus Ophiovirus
| Citrus psorosis virus |
|
|
| (Citrus ringspot virus) |
|
|
| Citrus psorosis virus P-121 | [AY654892-4=NC_006314-6] | (CPsV-P121) |
| Freesia sneak virus |
|
|
| (Freesia ophiovirus) |
|
|
| Freesia sneak virus-Fr220205/9 | [DQ885455*] | (FreSV-Fr220205/9) |
| Lettuce ring necrosis virus |
|
|
| Lettuce ring necrosis virus-Belg2 | [AY535016-9=NC_006051-4] | (LRNV-Belg2) |
| Mirafiori lettuce big-vein virus |
|
|
| (Mirafiori lettuce virus) |
|
|
| Mirafiori lettuce big-vein virus-LS301-O | [AF525933-6=NC_004779-82] | (MiLBVV-LS301-O) |
| Ranunculus white mottle virus |
|
|
| Ranunculus white mottle virus-Rn3 | [AF335429, AY542957*] | (RWMV-Rn3) |
| Tulip mild mottle mosaic virus |
|
|
| Tulip mild mottle mosaic virus-Bas-1 | [AY542958*] | (TMMMV-Bas-1) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.*Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Ophiovirus but have not been approved as species
None reported.
List of unassigned species in the family Ophioviridae
None.
Phylogenetic relationships within the family
Ophiovirus-specific primers, based on a highly conserved sequence of RNA-1, have been tested in RT-PCR with all ophiovirus species. In all cases, a 136 bp fragment was amplified. Phylogenetic analysis of the deduced 45 aa strings derived from the amplified sequences of available isolates for each species (Figure 3) supported the positions of CPsV, RWMV, LRNV and FreSV as distinct species and indicated a closer relationship between MiLBVV and TMMMV, as already suggested by serological tests and preliminary sequence data. CPsV appears to be more distantly related to the other members, not only from serological and molecular data but also because it infects a woody host and there is no evidence of soil-borne transmission. These differences, if reinforced, may also lead to the re-assignment of the existing species into two separate genera in the family.
Similarity with other taxa (Figure 4)
Ophiovirus virion morphology resembles that of the tenuiviruses and the internal nucleocapsid component of members of the family Bunyaviridae. Unlike tenuiviruses, ophioviruses do not infect plants in the Gramineae, and unlike members of the family Bunyaviridae they do not have enveloped virions. Moreover, ophioviruses do not have the conserved identical nucleotides at the genomic RNA termini that are typical of tenui- and phleboviruses (family Bunyaviridae). MiLBVV appears to have a terminal “corkscrew”-like conformation similar to that in Orthomyxoviridae. RdRp aa sequences show ophioviruses to be similar to members of the families Paramyxoviridae, Rhabdoviridae, Bornaviridae and Filoviridae, and the (unassigned) genus Varicosavirus (Figure 4). The RdRp also contains the SDD sequence in motif C, a signature for segmented negative stranded RNA viruses (families Orthomyxoviridae, Arenaviridae and Bunyaviridae). However, phylogenetic reconstructions using the sequences of the conserved RdRp motifs of representative negative stranded RNA viruses reinforce the taxonomic relatedness of the ophioviruses and suggest their separation as a monophyletic group.
Derivation of name
Ophio: from the Greek ophis, “serpent”, referring to the snaky appearance of the virions.
Further reading
Derrick et al., 1988 K.S. Derrick, R.H. Brlansky, J.V. Da Graça, R.F. Lee, L.W. Timmer, T.K. Nguyen, Partial characterization of a virus associated with citrus ringspot. Phytopathol. 78 (1988) 1298–1301.
Garcia et al., 1994 M.L. Garcia, E. Dal Bo, O. Grau, R.G. Milne, The closely related citrus ringspot and citrus psorosis viruses have particles of novel filamentous morphology. J. Gen. Virol. 75 (1994) 3585–3590.
Lot et al., 2002 H. Lot, R.N. Campbell, S. Souche, R.G. Milne, P. Roggero, Transmission by Olpidium brassicae of Mirafiori lettuce virus and Lettuce big-vein virus, and their roles in lettuce big-vein etiology. Phytopathol. 92 (2002) 288–293.
Morikawa et al., 1995 T. Morikawa, Y. Nomura, T. Yamamoto, T. Natsuaki, Partial characterization of virus-like particles associated with tulip mild mottle mosaic. Ann. Phytopathol. Soc. Jpn. 61 (1995) 578–581.
Roggero et al., 2000 P. Roggero, M. Ciuffo, A.M. Vaira, G.P. Accotto, V. Masenga, R.G. Milne, An ophiovirus isolated from lettuce with big-vein symptoms. Arch. Virol. 145 (2000) 2629–2634.
Vaira et al., 2003 A.M. Vaira, G.P. Accotto, A. Costantini, R.G. Milne, The partial sequence of RNA1 of the ophiovirus Ranunculus white mottle virus indicates its relationship to rhabdoviruses and provides candidate primers for an ophiovirus-specific RT-PCR test. Arch. Virol. 148 (2003) 1037–1050.
Vaira and Milne, 2008 A.M. Vaira, R.G. Milne, Ophiovirus. In: . Elsevier, Oxford2008447–454.
Van der Wilk et al., 2002 F. Van der Wilk, A.M. Dullemans, M. Verbeek, J.F.J.M. van den Heuvel, Nucleotide sequence and genomic organization of an ophiovirus associated with lettuce big-vein disease. J. Gen. Virol. 83 (2002) 2869–2877.
Contributed by
Vaira, A.M., Gago-Zachert, S., Garcia, M.L., Guerri, J., Hammond, J., Milne, R.G., Moreno, P., Morikawa, T., Natsuaki, T., Navarro, J.A., Pallas, V., Torok, V., Verbeek, M. and Vetten, H.J.
Figures
Figure 1 Negative contrast electron micrograph (uranyl acetate) of virions of an isolate of Mirafiori lettuce big-vein virus. The bar represents 100 nm.
(Courtesy of R. G. Milne.)
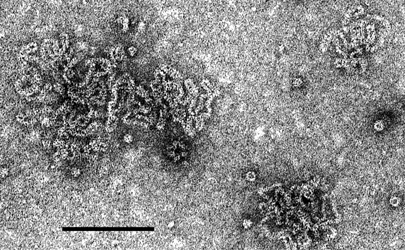
Figure 2 Genome organization of genus Ophiovirus. Mirafiori lettuce big-vein virus (MiLBVV) is shown here. Boxes represents ORFs. The length of the RNA segments and the predicted sizes of the ORF products are indicated. V, viral RNA; vc, viral complementary RNA. RNA-4 is not reported for all ophioviruses.
(Modified from van der Wilk et al., 2002.)
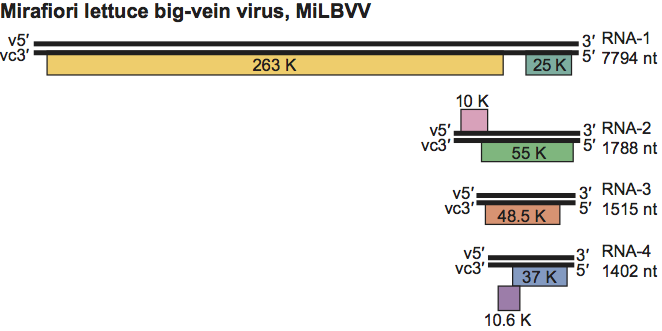
Figure 3 Unrooted phylogenetic tree based on the predicted amino acid sequences (45aa) encoded by the conserved 136 nt fragment of the RdRp gene, obtained from 20 ophiovirus isolates belonging to the six species. Alignment was obtained using ClustalW, and analyzed by the neighbor-joining method, with 10 000 bootstrap replications. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches (when >50%). The evolutionary distances were computed using the Poisson correction method.
(Courtesy of A. M. Vaira.)
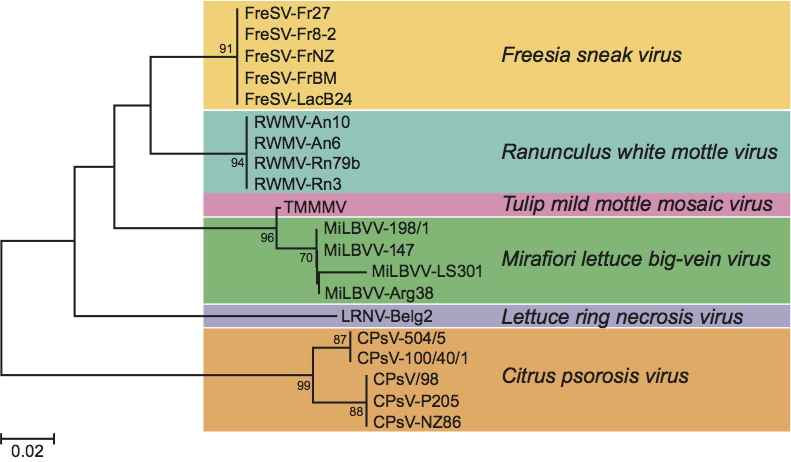
Figure 4 Unrooted phylogenetic tree of members of family Ophioviridae and other negative stranded RNA viruses based on their conserved RdRp modules. The tree was generated by the neighbor-joining method and bootstrap values (indicated for each branch node when >50%) were estimated using 1000 replicates. Viruses included in the analysis and the accession numbers used [ ] are: BDV [L27077], BEFV [AF234533], BUNV [X14383], CDV [NC_001921], CPsV [AY224663], DHOV [M65866], DUGV [U15018], FLUAV [J02151], FLUBV [M20170], FLUCV [M28060], HRSV [NC_001781], HTNV [X55901], IHNV [L40883], ISAV [AJ002475], LACV [U12396], LBVV [AB075039], LCMV [J04331], MARV [M92834], MEV [K01711], MiLV (MiLBVV) [AF525933], MuV [D10575], NCMV [NC_002251], NDV [X05399], NIV [AF212302], PUUV [M63194], RABV [M13215], RSV [D31879], RVFV [X56464], RWMV [AF335429], RYSV [NC_003746], SEOV [X56492], SeV [M19661], SNV [L37901], SYNV [L32603], TCRV [J04340], THOV [AF004985], TOSV [X68414], TRTV [U65312], TSWV [D10066], UUKV [D10759], VSIV [J02428], VSNJV [M20166], and ZEBOV [AF499101].
(Courtesy of S. Gago-Zachert and M. L. Garcia.)