Genus: Deltavirus
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions of hepatitis delta virus (HDV) are approximately spherical, with an average diameter of 36–43 nm and no visible surface projections (Figure 1). They consist of an outer envelope containing lipid and all three envelope proteins of the co-infecting helper hepadnavirus (human hepatitis B virus (HBV) in nature, though woodchuck hepatitis virus can also act as a helper in the laboratory) (see below), and an inner nucleocapsid of 19 nm comprising the RNA genome of HDV and approximately 70 copies of the only HDV-encoded protein, known as hepatitis delta antigen (HDAg). HDAg exists in two forms (large HDAg, L-HDAg or p27; and small HDAg, S-HDAg or p24), which differ only by a 19 amino acid residue C-terminal extension. Virions contain variable amounts of L-HDAg and S-HDAg in close association with virion RNA. Nucleocapsid symmetry has not been confirmed. Nucleocapsids can be released by treatment of virions with non-ionic detergent and dithiothreitol.
Physicochemical and physical properties
Virions have a buoyant density in CsCl of about 1.24 g cm−3. In rate zonal sedimentation they behave as smaller than HBV.
Nucleic acid
The genome consists of a single molecule of circular, negative sense ssRNA about 1.7 kb in length. With a high degree (ca. 70%) of intramolecular base pairing, it has the potential to fold on itself, forming an unbranched rod-like structure. Both genomic and antigenomic RNA species can function as a ribozyme to carry out self-cleavage and possibly self-ligation. The above properties make this genome unique and distinct from all other known animal viruses.
Proteins
HDV RNA encodes one known protein S-HDAg (see above). A second species, L-HDAg, arises via an RNA editing event mediated by a host double stranded RNA adenosine deaminase, ADAR1. As a result of its action, the UAG stop codon for S-HDAg is converted to UGG, thereby allowing readthrough translation giving rise to L-HDAg. Both HDAg species are multifunctional with domains identified that result in (from the N-terminus): (i) dimerization via a coiled coil structure; (ii) nuclear localization via a bipartite signal; (iii) RNA binding via two arginine-rich motifs. In addition, L-HDAg has a domain that includes a prenylation site required for packaging. The two HDAg species play distinct roles in replication; S-HDAg is essential for HDV replication while L-HDAg is essential for packaging and in some situations, may inhibit replication. HDAg can be phosphorylated at serine residues.
The remaining structural proteins of the HDV virion consist of the surface proteins and glycoproteins of the helper hepadnavirus located in the HDV envelope.
Lipids
These are present but have not been characterized.
Carbohydrates
Presumably these are only present on the envelope proteins of HBV, the natural helper virus.
Genome organization and replication
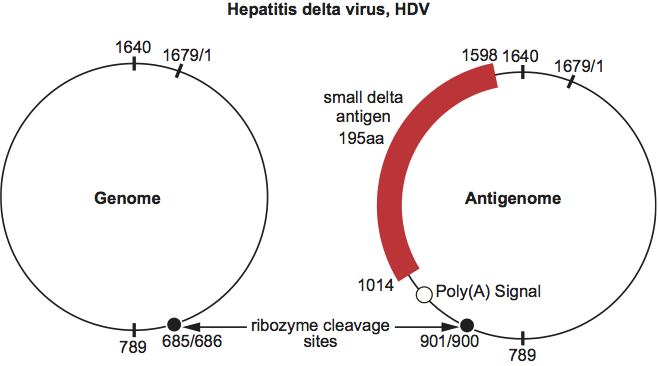
Attachment, entry and uncoating of HDV may be similar to the steps that occur with the helper hepadnavirus. Both viruses require the preS1 domain of the hepatitis B virus large envelope protein for attachment. Genome replication involves RNA-directed RNA synthesis carried out by host cell RNA polymerase II in the nucleus. An additional host polymerase might also be involved. Transcription is thought to occur by a double rolling circle mechanism that generates oligomeric forms of each complementarity, which then undergo site-specific autocatalytic cleavage and ligation to generate circular genomic and antigenomic monomers (Figure 2).
Only one HDV mRNA species has been identified, that coding for HDAg. In transfected cells, only S-HDAg is made initially and L-HDAg appears subsequently as a result of the RNA editing event described above. Such editing occurs during replication in tissue culture as well as in infected chimpanzees and woodchucks.
As HDV assembly requires the envelope proteins of a helper hepadnavirus, its assembly pathway is likely to overlap with that of the helper virus. In dually transfected cells, L-HDAg must be present for delta antigen-containing particles to be released, while S-HDAg is packaged if present in the cell, but is not sufficient for particle formation. Full size, or deleted, HDV RNA molecules are incorporated if present in the cell, as long as they are capable of folding into rod-like structures and binding with HDAgs. In cells undergoing HDV RNA replication, this process is highly specific for genomic RNA, while in cells expressing but not replicating HDV RNA, either sense can be assembled.
Antigenic properties
HDAg molecules have a unique antigenicity. Antibodies to these epitopes are diagnostic of current or past infections.
Biological properties
Full replication of HDV requires the presence of a helper hepadnavirus to provide envelope proteins, and HDV can therefore be considered as a subviral satellite virus. Natural HDV infection is found only in humans with HBV as helper virus. However, it can be transmitted to chimpanzees if accompanied by HBV, and experimental transmission of HDV to woodchucks has also been achieved using woodchuck hepatitis virus as helper virus.
Transmission of HDV to laboratory mice has been reported, leading to a single round of HDV genome replication in hepatocytes but no further replication, presumably due to the absence of helper virus.
Transmission of HDV in humans occurs by similar routes to those utilized by HBV, although, in many parts of the world, transmission by parenteral contact (e.g. sharing of intravenous needles) is more prominent than sexual or vertical routes. If transmission occurs to an individual with chronic HBV infection, this situation is termed super-infection and HDV infection usually then persists. On the other hand, if both HDV and HBV are simultaneously transmitted to a naïve host, the situation is termed co-infection and both infections are usually transient. HDV distribution is world-wide, but the proportion of HBV carriers who also have chronic HDV infection varies greatly between 0% and 60% in different geographical areas.
Clinical sequelae of acute and chronic HDV infection are variable and cover a similar spectrum to those of HBV alone. They include acute hepatitis, chronic active hepatitis, cirrhosis, fulminant acute hepatitis and hepatocellular carcinoma. However, the frequency of severe sequelae and their rates of progression are significantly higher in chronic HDV infection than in chronic HBV infection alone. A subacute, rapidly progressive form of HDV super-infection has been seen in HBV carriers in Venezuela, and other forms of severe acute and chronic hepatitis D, often fatal, occur in indigenous populations of Venezuela, Colombia, Brazil and Peru.
Species demarcation criteria in the genus
Humans are the only known natural host for HDV. Only one species is identified in the genus. Sequencing independent isolates of HDV revealed up to 40% variation in nucleotide sequence with up to 25 nucleotides variation in length. Studies of nucleotide homologies have distinguished eight major monophyletic groups or genotypes, HDV1 to HDV8. There is some geographical clustering, with HDV-1 found in USA, Europe and China, HDV-2 and HDV-4 in Japan and Taiwan, HDV-3 in South America predominantly associated with HBV/F, and HDV-5 to HDV-8 in Africa.
List of species in the genus Deltavirus
| Hepatitis delta virus |
|
|
| Hepatitis delta virus - 1 (USA, Europe, China) | [X04451] | (HDV-1) |
| Hepatitis delta virus - 2 (Japan) | [X60193] | (HDV-2) |
| Hepatitis delta virus - 3 (South America) | [L22063] | (HDV-3) |
| Hepatitis delta virus - 4 (Taiwan, Japan) | [AF018077] | (HDV-4) |
| Hepatitis delta virus - 5 (Africa) | [AJ584848] | (HDV-5) |
| Hepatitis delta virus - 6 (Africa) | [AJ584847] | (HDV-6) |
| Hepatitis delta virus - 7 (Africa) | [AJ584844] | (HDV-7) |
| Hepatitis delta virus - 8 (Africa) | [AJ584849] | (HDV-8) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses that may be members of the genus Deltavirus but have not been approved as species
None reported.
Similarity with other taxa
Several features of HDV (genome structure, RNA-RNA transcription using RNA polymerase II, and autocatalytic RNA sites) are similar to properties of some viroids. However, unlike viroids, HDV possesses a larger genome, encodes a functional protein and requires a specific hepadnavirus helper function.
HDV also possesses some features in common with ssRNA satellites of plants, including both the large and circular ssRNA satellites and also (in terms of the satellite-helper relationship) with ssRNA satellite viruses such as chronic bee-paralysis virus-associated satellite virus and tobacco necrosis satellite virus.
However, on the basis of genome size and structure, mode of replication, protein coding strategy, structure of virion and satellite-helper virus relationship, none of the above examples warrants inclusion in a distinct family together with HDV.
Derivation of name
Delta: A novel antigen in HBV infected tissue, unrelated to previously described HBV antigens, was named delta antigen (δAg) by Rizzetto et al. in 1977.
Further reading
Deny, 2006 P. Deny, J.L. Casey, Hepatitis delta virus genetic variability: from genotype I, II III to eight major cladesHepatitis Delta Virus. In: J.L. Casey, Hepatitis Delta Virus. Springer Verlag, Heidelberg2006151–157.
Rizzetto et al., 1977 M. Rizzetto, M.G. Canese, S. Arico, O. Crivelli, F. Bonino, C.G. Trepo, G. Verme, Immunofluorescence detection of a new antigen-antibody system (δ/anti-δ) associated to hepatitis B virus in liver and serum of HBsAg carriers. Gut. 18 (1977) 977–1003.
Taylor et al., 2006 J.M. Taylor, P. Farci, R.H. Purcell, D.M. Knipe, P.M. Howley, Hepatitis D (delta) virusFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott Williams & Wilkins, Philadelphia20063031–3046.
Contributed by
Mason, W.S., Gerlich, W.H., Taylor, J.M., Kann, M., Mizokami, M., Loeb, D., Sureau, C., Magnius, L. and Norder, H.
Figures
Figure 1 Schematic representation of a particle of hepatitis delta virus.
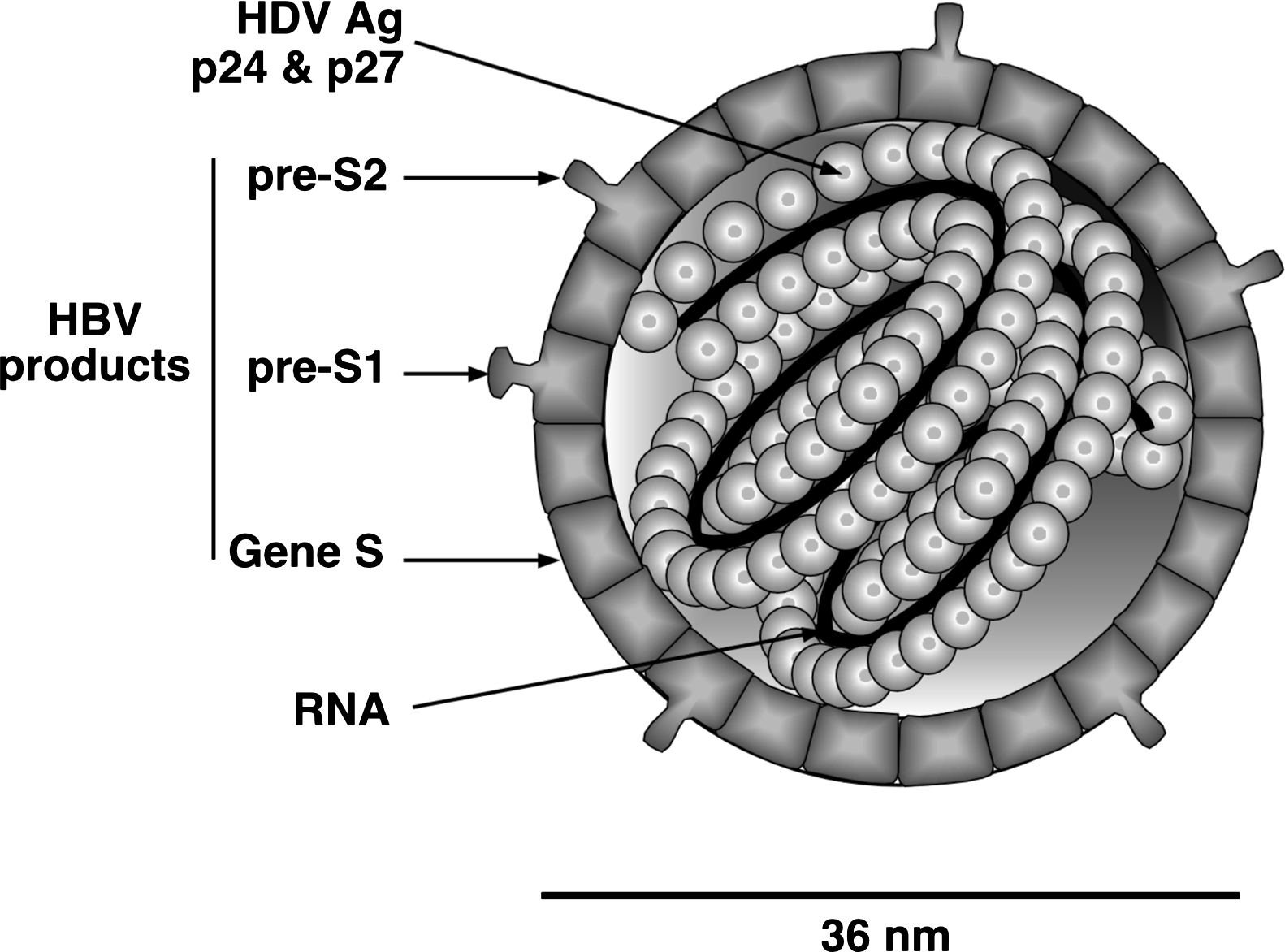
Figure 2 Organization of the genome and antigenome of hepatitis delta virus. The genome, by definition, is the 1679-nucleotide RNA that is assembled into virions. During replication one also detects the antigenome, a species that is an exact complement of the genome. Both RNAs are circular in conformation and have the ability to fold into an unbranched rodlike structure via intra-molecular base pairing, with the locations of the ends as indicated. Each has a ribozyme, with the cleavage sites as indicated. The antigenome contains the ORF for the 195 amino acid small delta antigen. However, this protein is actually translated from a third RNA, one that is only about 900 nt long, linear in conformation, 5-capped and 3-polyadenylated.