Family: Arenaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Arenavirus
Type species Lymphocytic choriomeningitis virus
Virion properties
Morphology
Virions are spherical to pleomorphic, 50–300 nm in diameter (mean 110–130 nm), with a dense lipid envelope and a surface layer covered by club-shaped projections, 8–10 nm in length. A variable number of 20–25 nm ribosomes are generally present within virus particles. Isolated nucleocapsids, free of contaminating host ribosomes, are organized in closed circles of varying length (450–1300 nm), which have been shown to assume supercoiled forms, and display a linear array of nucleosomal subunits.
(Left) Electron microscopic images of lymphocytic choriomeningitis virus (LCMV). (A) Thin section showing several virions budding from the surface of an infected BHK-21 cell. (B) Cryo-electron microscopic images of purified unstained virions frozen in vitreous ice. Arrowheads indicate glycoprotein spikes which are composed of trans-membrane GP2 and globular heads of GP1. The bar indicates 100 nm. (Courtesy R. Milligan, J. Burns and M. Buchmeier). (C) Diagrammatic representation of virion structure with trimeric spikes (Eschli et al., 2006; Schlie et al., 2010). L protein is the RNA polymerase; IB is inclusion bodies that could be ribosomes or could be related to self-assembling Z bodies (Kentsis et al., 2002).
(Courtesy C. Clegg and A. Featherstone, [email protected].)
Physicochemical and physical properties
Virion Mr has not been determined. The S20,w is 325–500. The buoyant density in sucrose is about 1.17–1.18 g cm−3, in CsCl it is about 1.19–1.20 g cm−3, in amidotrizoate compounds it is about 1.14 g cm−3. Virions are relatively unstable in vitro, and are rapidly inactivated below pH 5.5 and above pH 8.5. Virus infectivity is inactivated at 56 °C, by treatment with organic solvents or detergents, or by exposure to UV- and gamma-irradiation.
Nucleic acid
The genome consists of two single stranded, ambisense RNA molecules, L and S, of lengths of about 7.5 kb and 3.5 kb, respectively. There are no poly(A) tracts at the 3′ termini. The 3′-terminal sequences (19–30 nucleotides) are similar in the two RNAs and among different arenaviruses. Overall, they are largely complementary to the 5′-end sequences. Although the RNA genomic species are thought to be present in virions in the form of circular nucleocapsids, the genomic RNA is not covalently closed. Variable amounts of full-length viral-complementary RNAs (predominantly S) and viral subgenomic mRNA species have been reported in virus preparations. Preparations of purified virus may also contain RNAs of cellular origin with sedimentation coefficients of 28S, 18S and 4–6S. These include ribosomal RNAs. The viral mRNA species may be associated with encapsidated ribosomes, though another possibility is that the dense inclusion bodies seen in virions are related to self-assembling Z bodies. The RNA species are not present in equimolar amounts, apparently due to the packaging of multiple RNA molecules per virion. For example, virions may package more than one S RNA molecule, as well as RNA molecules with complementary sequences.
Proteins
The most abundant structural protein is the nucleoprotein (N or NP), a non-glycosylated polypeptide (ca. 63 kDa) found tightly associated with the virus genomic RNA in the form of a ribonucleoprotein complex or nucleocapsid structure. A minor component is the L protein, an RNA polymerase (ca. 200 kDa). A zinc binding protein (Z or p11; 10–14 kDa) is also a structural component of the virus, and functions as a matrix protein (with roles in virus assembly/disassembly and inhibiting transcription). Two glycosylated proteins (GP1 or G1, GP2 or G2; 34–44 kDa) are found in all members of the family and are derived by posttranslational cleavage from an intracellular precursor, GPC (ca. 75–76 kDa). A stable signal peptide (SSP) cleaved during GPC synthesis is also in the virion spike. Other minor proteins and enzymatic activities have been described associated with virions including poly (U) and poly (A) polymerases, and a protein kinase that can phosphorylate N. It is thought unlikely that these are virally encoded.
Lipids
Lipids represent about 20% of virion dry weight and are similar in composition to those of the host plasma membrane.
Carbohydrates
Carbohydrates in the form of complex glycans on GP1 (five or six sites in lymphocytic choriomeningitis virus [LCMV]) and GP2 (2 sites in LCMV) represent about 8% of virion dry weight.
Genome organization and replication
The L and S RNAs of arenaviruses each have an ambisense coding arrangement (Figure 2). The L RNA encodes in its viral-complementary sequence the L protein, and in the viral-sense 5′-end sequence the Z protein. The Z mRNA is small (<0.5 kb). The N protein is encoded in the viral-complementary sequence corresponding to the 3-half of the S RNA, while the viral glycoprotein precursor (GPC) is encoded in the viral-sense sequence corresponding to the 5′-half of the S RNA. The two proteins are made from subgenomic mRNA species transcribed from the viral (for N mRNA) or full-length viral-complementary S RNA species (for GPC mRNA). The intergenic regions of both S and L RNAs contain nt sequences with the potential of forming one or more hairpin configurations. These secondary structural features may function to terminate mRNA transcription from the viral and viral-complementary S RNAs. The mRNAs are capped and contain 1-5 non-templated nt of heterogeneous sequence at their 5′ ends. The mRNAs are not polyadenylated. The transcription mechanism is not fully elucidated. Initiation of transcription may involve cap-snatching. The 3′ termini of the mRNAs have been mapped to locations in the intergenic regions.
The process of infection involves attachment to cell receptors, entry via the endosomal route, uncoating and mRNA transcription in the cytoplasm of infected cells. Because of the ambisense coding arrangement, only N and L mRNAs can be synthesized from the genomic RNAs by the virion polymerase prior to translation. The products of these mRNAs are presumed to be involved in the synthesis of full-length viral complementary species which serve as templates for the synthesis of GPC and Z mRNAs and the synthesis of full-length viral RNAs. The process of RNA replication, which may involve a slippage mechanism during initiation, and readthrough of transcription termination signals, has not been fully elucidated. However, the presence of full-length viral-complementary genomic RNAs and viral sgRNA species in virus preparations may affect this perceived temporal order of RNA and protein synthesis.
The viral envelope glycoproteins are synthesized in cells as a single mannose-rich precursor molecule that is proteolytically cleaved and processed to contain complex glycans during transport to the plasma membrane. First, its signal peptide is co-translationally cleaved and mediates the cleavage of GPC into GP1 and GP2 by SKI protease as well as the assembly of virions. Virions mature by budding at sites on the surface of cells. Ribosomes are also observed at such sites. Interstrain reassortant progeny can be formed, including diploid (or multiploid) species with respect to the genomic RNA segments. Evidence for interspecies reassortment between Lassa virus (LASV) and Mopeia virus has also been obtained. Ribavirin inhibits the replication of several arenaviruses in vitro and is effective in the therapy of humans and primates infected with Lassa virus at early disease stage. Several antiviral agents targeting different stages of viral replication are under evaluation.
Antigenic properties
Viruses possess a number of distinct antigenic determinants as shown by monoclonal and polyclonal antibody analyses. Antigens on the 44 kDa GP1 of lymphocytic choriomeningitis virus (LCMV) are involved in virus neutralization. These are type-specific, although cross-neutralization tests have demonstrated partially shared antigens between Tacaribe virus and Junín virus. Cross-protection has also been demonstrated against Junín virus following prior infection by Tacaribe virus, or against Lassa virus following infection by Mopeia virus. Major complement-fixing antigens are associated with the viral N proteins, which were used to define the Tacaribe complex of arenaviruses. Monoclonal antibodies react with common epitopes on the N proteins of all arenaviruses and a single highly conserved epitope has also been described in the transmembrane GP2 glycoprotein.
By analyses using monoclonal and polyclonal antibody, the African arenaviruses are distinguishable from the New World arenaviruses. Fluorescent antibody studies show that antisera against New World viruses, as well as those against African viruses, react with LCMV. Cytotoxic T-lymphocyte epitopes have been identified on the nucleoprotein and glycoproteins of LCMV. The number and location of epitopes varies depending on the virus strain and host MHC class I molecules. No hemagglutinin has been identified.
Biological properties
The reservoir hosts of almost all the arenaviruses are species of rodents. LCMV is found in mouse and the African viruses mainly in the rodents Mastomys and Praomys, in the sub-family Murinae. The New World viruses are mostly found in the Sigmodontine rodents Calomys, Neacomys, Neotoma, Oryzomys and Sigmodon. Exceptionally, Tacaribe virus was isolated from fruit-eating bats (Artibeus spp.), but subsequent attempts to recover it from bats or from other potential hosts have been unsuccessful. It is notable that the geographic range of an arenavirus is generally much more restricted than that of its cognate rodent host. Most of the viruses induce a persistent, frequently asymptomatic infection in their reservoir hosts, in which chronic viremia and viruria occur. Such infections are known or suspected to be caused by a slow and/or insufficient host immune response. Most arenaviruses do not normally infect other mammals or humans. However, Lassa virus is the cause of widespread human infection (Lassa fever) in West Africa (Nigeria, Sierra Leone, Liberia, Guinea), LuJo virus has caused a Lassa fever-like outbreak in South Africa, and Junín virus causes Argentine hemorrhagic fever in agricultural workers in an increasingly large area of that country. Machupo virus (MACV) has caused isolated outbreaks of similar disease in Bolivia, and Guanarito virus (GTOV) is associated with human disease in Venezuela. Sabiá virus was isolated from a fatal human case in Brazil and a related virus, Chapare was isolated in Bolivia. Human infection with LCMV may occur in some rural and urban areas with high rodent populations, and has been acquired from pet hamsters. Organ transplants from LCMV-infected individuals have resulted in at least 10 human deaths since 1998. LCMV acquired from mice has also caused a highly fatal hepatitis in captive Callitrichid primates. Severe laboratory-acquired infections have occurred with LCMV, Lassa, Junín, Machupo, Sabiá and Flexal viruses. Asymptomatic infections with Pichinde virus have been reported.
Success of experimental infection in laboratory animals (mouse, hamster, guinea pig, rhesus monkey, marmoset, rat) varies with the animal species and the virus. In general, New World viruses are pathogenic for suckling but not weaned mice; LCMV and Lassa virus produce the opposite effect. Viruses grow moderately well in many mammalian cells. Receptors mediating host cell entry are thought to be alpha-dystroglycan for Old World viruses (e.g. some strains of LCMV and LASV) and Clade C New World viruses, and transferrin receptor-1 for New World viruses (MACV, GTOV, Junín virus [JUNV]); however virus entry by some strains has been observed in the absence of either receptor. Arenaviruses primarily infect cells of the myeloid and reticuloendothelial lineages but are also found in hepatocytes, lymphocytes and other cells.
Vertical and horizontal (including venereal) transmissions occur in the natural hosts. These include transuterine, transovarian and post-partum transmission and can be via milk-, saliva- or urine-borne routes. Horizontal transmission within and between host species occurs by contamination and aerosol routes. No arthropod vectors are thought to be involved in the normal transmission process.
Species demarcation criteria in the genus
The parameters used to define a species in the genus are:
- an association with a specific host species or group of species;
- presence in a defined geographical area;
- etiological agent (or not) of disease in humans;
- significant differences in antigenic cross-reactivity, including lack of cross-neutralization activity where applicable;
- significant amino acid sequence difference from other species in the genus (i.e. showing a divergence between species of at least 12% in the nucleoprotein amino acid sequence).
For example, although both Pirital virus and Guanarito virus circulate in the same region of Venezuela, they are distinguished by their isolation from different rodent hosts (Sigmodon alstoni and Zygodontomys brevicauda, respectively). In addition, in ELISA with hyperimmune mouse ascitic fluids, titers differ by at least 64-fold, and sequence analysis shows less than 55% aa identity between partial nucleocapsid protein sequences. In another example, both Lassa virus and Mopeia virus share a common rodent host (Mastomys) at the genus level. However, they are distinguished by their different geographical range, different profiles of reactivity with panels of monoclonal antibodies, and by N protein aa sequence divergences of about 26%. Also, Lassa virus is the cause of hemorrhagic fever in humans and other primates, while Mopeia virus is not associated with human disease and does not cause disease in experimentally infected primates.
List of species in the genus Arenavirus
| Old World arenaviruses |
|
|
| Ippy virus |
|
|
| Ippy virus - Dak AN B 188d | [S segment: DQ328877, L segment: DQ328878] | (IPPYV) |
| Arvicanthis sp., Central African Republic |
|
|
| Lassa virus |
|
|
| Lassa virus - GA391 | [S segment: X52400, L segment: U73034] | (LASV-GA391) |
| Mastomys sp., West Africa |
|
|
| Lymphocytic choriomeningitis virus |
|
|
| Lymphocytic choriomeningitis virus - Armstrong 53b | [S segment: AY847350 | (LCMV-Ar53b) |
| Mus musculus, Europe, Americas | L segment: AY847351] |
|
| Mobala virus |
|
|
| Mobala virus - 3080 | [S segment: AY342390, L segment: DQ328876] | (MOBV-3076) |
| Praomys sp., Central African Republic |
|
|
| Mopeia virus |
|
|
| Mopeia virus - AN 20410 | [S segment: AY772170, L segment: AY772169] | (MOPV-AN20410) |
| Mastomys natalensis, Mozambique, Zimbabwe |
|
|
| Morogoro virus | [S segment: EU914103, L segment: EU914104] | (MORV) |
| Mastomys natalensis, Zambia |
|
|
| New World arenaviruses |
|
|
| Allpahuayo virus |
|
|
| Allpahuayo virus - CLHP-2472 | [S segment: AY012687, L segment: AY216502] | (ALLV-CLHP2472) |
| Oecomys bicolor, Oe. paricola |
|
|
| Amapari virus |
|
|
| Amapari virus - BeAn 70563 | [S segment: AF485256, L segment: AY216517] | (AMAV-BeAn70563) |
| Oryzomys capito, Neacomys guianae, Brazil |
|
|
| Bear Canyon virus |
|
|
| Bear Canyon virus - A0070039 | [S segment: AY924391, L segment: AY924390] | (BCNV-A0060209) |
| Peromyscus californicus |
|
|
| Chapare virus |
|
|
| Chapare virus - 810419 | [S segment EU260463, L segment EU260464] | (CHPV-810419) |
| Homo sapiens, Bolivia |
|
|
| Cupixi virus |
|
|
| Cupixi virus - BeAn 119303 | [S segment: AF512832, L segment: AY216519] | (CPXV-BeAn 119303) |
| Oryzomys sp. |
|
|
| Flexal virus |
|
|
| Flexal virus - BeAn 293022 | [S segment: AF512831, L segment: EU627611] | (FLEV-BeAn293022) |
| Oryzomys spp., Brazil |
|
|
| Guanarito virus |
|
|
| Guanarito virus - INH-95551 | [S segment: AY129247, L segment: AY358024] | (GTOV-INH95551) |
| Zygodontomys brevicauda, Venezuela |
|
|
| Junín virus |
|
|
| Junín virus - XJ13 | [S segment: AY358023, L segment: AY358022] | (JUNV-XJ13) |
| Calomys musculinus, Argentina |
|
|
| Latino virus |
|
|
| Latino virus - 10924 | [S segment: AF485259, L segment: EU627612] | (LATV-10924) |
| Calomys callosus, Bolivia |
|
|
| Machupo virus |
|
|
| Machupo virus - Carvallo | [S segment: AY129248, L segment: AY358021] | (MACV-Carvallo) |
| Calomys callosus, Bolivia |
|
|
| Oliveros virus |
|
|
| Oliveros virus - RIID 3229 | [S segment: U34248, L segment: AY216514] | (OLVV-RIID3229) |
| Bolomys obscurus, Argentina |
|
|
| Paraná virus |
|
|
| Paraná virus - 12056 | [S segment: AF485261, L segment: EU627613] | (PARV-12056) |
| Oryzomys buccinatus, Paraguay |
|
|
| Pichinde virus |
|
|
| Pichinde virus - 3739 | [S segment: K02734, L segment: AF427517] | (PICV-3739) |
| Oryzomys albigularis, Colombia |
|
|
| Pirital virus |
|
|
| Pirital virus - VAV488 | [S segment: AF485262, L segment: AY494081] | (PIRV-VAV488) |
| Sigmodon alstoni, Venezuela |
|
|
| Sabiá virus |
|
|
| Sabiá virus - SPH114202 | [S segment: U41071, L segment: AY358026] | (SABV-SPH114202) |
| Natural host unknown, Brazil |
|
|
| Tacaribe virus |
|
|
| Tacaribe virus - p2b2 | [S segment: M20304 ] | (TCRV-p2b2) |
| Artibeus spp., Trinidad |
|
|
| Tacaribe virus - TRVLII573 | [L segment: J04340] | (TCRV-TRVLII573) |
| Artibeus spp., Trinidad |
|
|
| Tamiami virus |
|
|
| Tamiami virus - W10777 | [S segment: AF485263, L segment: AY924393] | (TAMV-W10777) |
| Sigmodon hispidus, Florida, USA |
|
|
| Whitewater Arroyo virus |
|
|
| Whitewater Arroyo virus – AV 9310135 | [S segment: AF228063, L segment: AY924395] | (WWAV-AV9310135) |
| Neotoma albigula, New Mexico, USA |
|
|
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Arenavirus but have not been approved as species
| Lujo virus(Lusaka/Johannesburg virus) | [S segment: FJ952384, L segment: FJ952385] | (LUJV) |
| Kodoko virusMus minutoides/Africa | [N gene: EF189586*, L segment: EF179864*] | (KODV-TA777, KD42) |
| Dandenong virusFound in an Australian transplant recipient of an organ from an eastern European donor | [S segment: EU136038, L segment: EU136039] | (DANV) |
| Merino Walk virus Myotomys unisulcatus/S Africa | [S segment: GU078660, L segment: GU078661] | (MWV) |
* Partial sequences.
Phylogenetic relationships within the family Arenaviridae
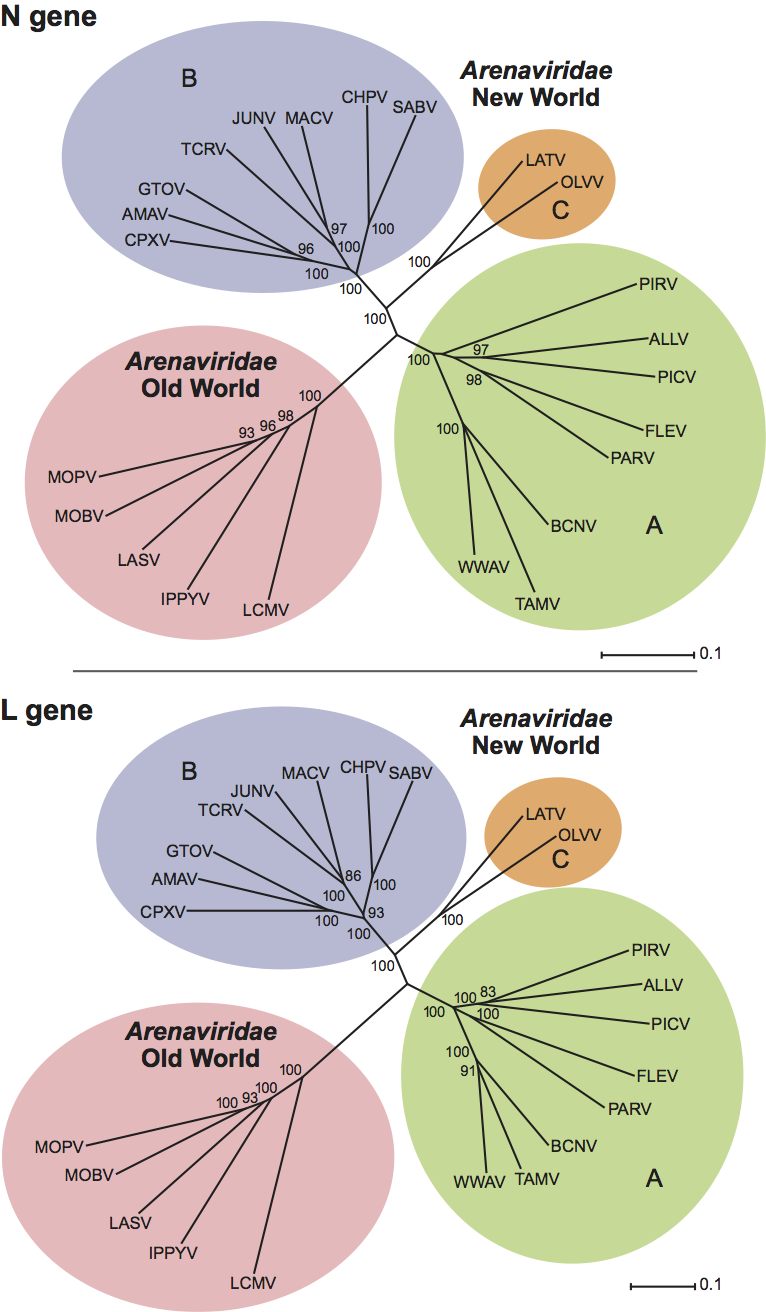
Nucleic acid sequences from the N genes of all the known arenaviruses have provided the basis for phylogenetic analysis that supports previously defined antigenic groupings and further defines virus relationships within them. Sequence data derived from other regions of the genome including the L gene are largely consistent with this analysis (Figure 3). Among the Old World viruses, Lassa virus, Mopeia virus and Mobala virus are monophyletic, while Ippy virus and lymphocytic choriomeningitis virus are more distantly related. One interesting virus, LuJo virus, found in South Africa, is most related to Old World viruses but contains elements of New World sequence in its glycoprotein. The New World viruses can be divided into three groups on the basis of the sequence data. In group A are Pirital virus, Pichinde virus, Paraná virus, Flexal virus, and Allpahuayo (Peru) virus from South America, together with Tamiami virus, Whitewater Arroyo virus and Bear Canyon virus from North America. Group B contains the human pathogenic viruses Machupo virus, Junín virus, Guanarito virus, Sabiá virus and Chapare virus as well as the non-pathogenic Tacaribe virus, Amapari virus, and Cupixi virus (from Brazil). Latino virus and Oliveros virus form a small separate group (group C). The division of the arenaviruses into Old World and New World groups, as well as the subdivision of New World arenaviruses into three groups, is strongly supported by bootstrap resampling analysis. It is important to note that the trait of human pathogenicity appears to have arisen on at least two independent occasions during arenavirus evolution.
Recombination may have influenced the evolution of arenaviruses. The nucleocapsid and glycoprotein genes of Whitewater Arroyo virus, Tamiami virus, and the Bear Canyon virus have divergent phylogenetic histories. Separate analysis of full-length amino acid sequences using maximum parsimony or neighbor-joining methods show that the nucleocapsid protein genes of these three viruses are related to those of Pichinde virus and Pirital virus (New World lineage A), while the glycoprotein genes are more closely related to those of Junín virus, Tacaribe virus, and Sabia virus (New World lineage B).
Similarity with other taxa
The Arenaviridae are unique amongst the negative strand viruses for their bi-segmented genome with ambisense coding strategy. Arenaviruses are most similar to the Bunyaviridae, another segmented negative-strand RNA virus family, whose members have three genome segments, some of which also encode genes in both senses.
Derivation of name
Arena: from Latin arenosus, “sandy” and arena, “sand”, in recognition of the sand-like particles observed in thin section. The name originally proposed was arenovirus, but was subsequently changed to avoid possible confusion with adenovirus.
Further reading
Archer and Rico-Hesse, 2002 A.M. Archer, R. Rico-Hesse, High genetic divergence and recombination in Arenaviruses. Virology. 304 (2002) 274–281.
Bowen et al., 1997 M.D. Bowen, C.J. Peters, S.T. Nichol, Phylogenetic analysis of the Arenaviridae: patterns of virus evolution and evidence for cospeciation between arenaviruses and their rodent hosts. Mol. Phylogenet. Evol. 8 (1997) 301–316.
Briese et al., 2009 T. Briese, J.T. Paweska, L.K. McMullan, S.K. Hutchison, C. Street, G. Palacios, M.L. Khristova, J. Weyer, R. Swanepoel, M. Egholm, S.T. Nichol, W.I. Lipkin, Genetic detection and caracterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from Southern Africa. PLoS Pathog. 4 (2009) e1000455.
Charrel et al., 2001 R.N. Charrel, X. de Lamballerie, C.F. Fullhorst, The Whitewater Arroyo virus: natural evidence for genetic recombination among Tacaribe serocomplex viruses (family Arenaviridae). Virology. 283 (2001) 161–166.
Charrel et al., 2002 R.N. Charrel, H. Feldmann, C.F. Fulhorst, R. Khelifa, R. de Chesse, X. de Lamballerie, Phylogeny of New World arenaviruses based on the complete coding sequences of the small genomic segment identified an evolutionary lineage produced by intra-segmental recombination. Biochem. Biophys. Res. Commun. 296 (2002) 1118–1124.
Delgado et al., 2008 S. Delgado, B.R. Erickson, R. Agudo, P.J. Blair, E. Vallejo, C.G. Albarino, J. Vargas, J.A. Comer, P.E. Rollin, T.G. Ksiazek, J.G. Olson, S.T. Nichol, Chapare virus, a newly discovered arenavirus isolated from a fatal hemorrhagic fever case in Bolivia. PLoS Pathog. 4 (6) (2008) e1000047.
Eschli et al., 2006 B. Eschli, K. Quirin, A. Wepf, J. Weber, R. Zinkernagel, H. Hengartner, Identification of an N-terminal trimeric coiled-coil core within arenavirus glycoprotein 2 permits assignment to class I viral fusion proteins. J. Virol. 80 (2006) 5897–5907.
Fulhorst et al., 2002 C.F. Fulhorst, S.G. Bennett, M.L. Milazzo, H.L. Murray, J.P. Webb, M.N.B. Cajimat, R.B. Bradley, Bear canyon virus: an arenavirus naturally associated with the California mouse (Peromyscus californicus). Emerg. Infect. Dis. 8 (2002) 717–721.
Kentsis et al., 2002 A. Kentsis, R.E. Gordon, K.L. Borden, Control of biochemical reactions through supramolecular RING domain self-assembly. Proc. Natl Acad. Sci., U S A. 99 (2002) 15404–15409.
Oldstone, 2002 Oldstone, M.B.A. (ed.) (2002). Arenaviruses I. Curr. Top. Microbiol. Immunol., 262, 1–197.
Oldstone, 2002 Oldstone, M.B.A. (ed.) (2002). Arenaviruses II. Curr. Top. Microbiol. Immunol., 263, 1–268.
Peters et al., 1996 C.J. Peters, M. Buchmeier, P.E. Rollin, T.G. Ksiazek, B.N. Fields, D.M. Knipe, P.M. Howley, ArenavirusesFields Virology. In: B.N. Fields, D.M. Knipe, P.M. Howley, Fields Virology. Lippincott-Raven, Philadelphia19961521–1552.
Radoshitzky et al., 2008 S.R. Radoshitzky, J.H. Kuhn, C.F. Spiropoulou, C.G. Albarino, D.P. Nguyen, J. Salazar-Bravo, T. Dorfman, A.S. Lee, E. Wang, S.R. Ross, H. Choe, M. Farzan, Receptor determinants of zoonotic transmission of New World hemorrhagic fever arenaviruses. Proc. Natl Acad. Sci., U S A. 105 (2008) 2664–2669.
Salvato, 1993 M.S. Salvato, The Arenaviridae. In: M.S. Salvato, The Arenaviridae. Plenum Press, New York1993.
Schlie et al., 2010 K. Schlie, A. Maisa, F. Lennartz, U. Stroher, W. Garten, T. Strecker, Characterization of Lassa virus glycoprotein oligomerization and influence of cholesterol on virus replication. J. Virol. 84 (2010) 983–992.
Contributed by
Salvato, M.S., Clegg, J.C.S., Buchmeier, M.J., Charrel, R.N., Gonzalez, J.P., Lukashevich, I.S., Peters, C.J. and Romanowski, V.
Figures
Figure 1. (Left) Electron microscopic images of lymphocytic choriomeningitis virus (LCMV). (A) Thin section showing several virions budding from the surface of an infected BHK-21 cell. (B) Cryo-electron microscopic images of purified unstained virions frozen in vitreous ice. Arrowheads indicate glycoprotein spikes which are composed of trans-membrane GP2 and globular heads of GP1. The bar indicates 100 nm. (Courtesy R. Milligan, J. Burns and M. Buchmeier). (C) Diagrammatic representation of virion structure with trimeric spikes (Eschli et al., 2006; Schlie et al., 2010). L protein is the RNA polymerase; IB is inclusion bodies that could be ribosomes or could be related to self-assembling Z bodies (Kentsis et al., 2002).
(Courtesy C. Clegg and A. Featherstone, [email protected].)
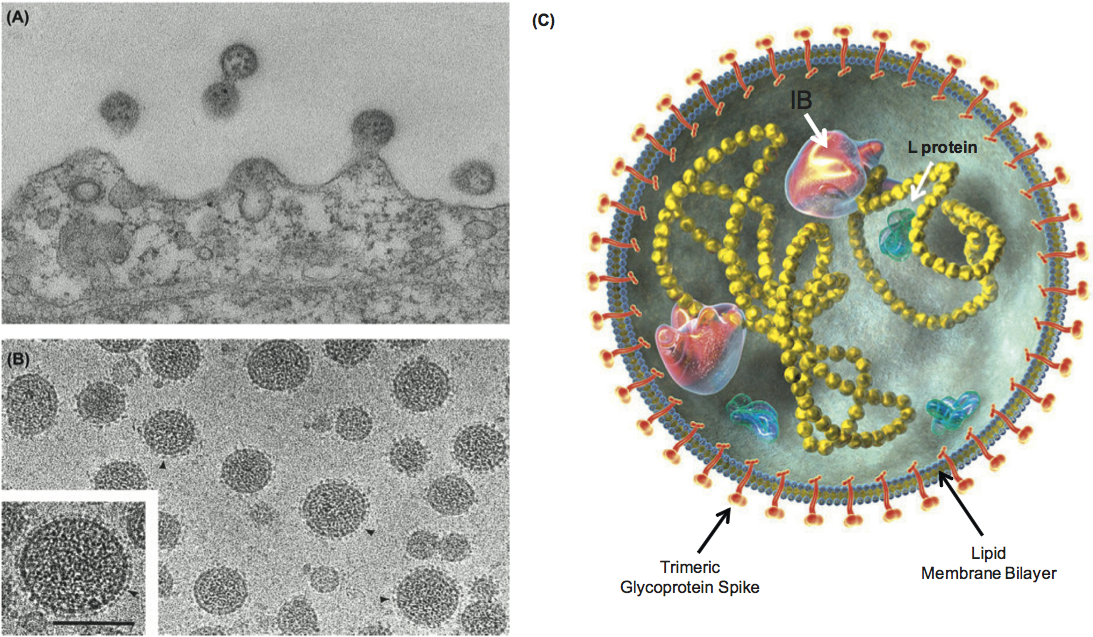
Figure 2 Organization, transcription and replication of the arenavirus L and S RNAs. Regions encoding the L, Z, GPC and N proteins are shown as boxes with arrowheads indicating the notional direction of translation. The intergenic regions separating the ORFs are indicated by gray boxes. RNA transcription processes are indicated by solid arrows.
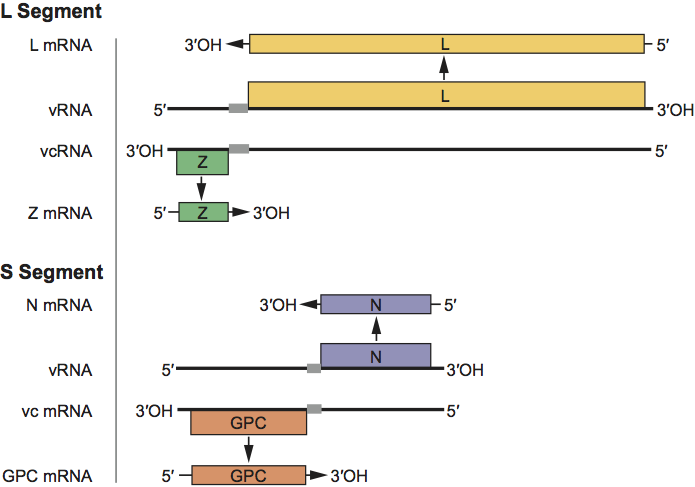
Figure 3 Phylogenetic relationships among the Arenaviridae. N and L gene codon-aligned nt sequences of an isolate of each species (Table 1) were analyzed in MEGA 4 (maximum composite likelihood distances and 10,000 bootstrap replicates). Bootstrap values are shown at the branches if >80%.