Family: Plasmaviridae
Mart Krupovic
The citation for this ICTV Report chapter is the summary published as Krupovic et al., (2018):
ICTV Virus Taxonomy Profile: Plasmaviridae, Journal of General Virology, 99: 617–618.
Corresponding author: Mart Krupovic ([email protected])
Edited by: Andrew M. Kropinski and Stuart G. Siddell
Posted: March 2018
PDF: ICTV_Plasmaviridae.pdf
Summary
The family Plasmaviridae includes bacterial viruses with slightly pleomorphic, enveloped virions with a diameter of 50–125 nm (Table 1. Plasmaviridae). Virions contain infectious, circular, supercoiled double-stranded DNA molecule(s) of ~12 kbp. Plasmaviruses infect Acholeplasma species, wall-less bacteria of the class Mollicutes, and are released by budding through the cell membrane without causing host cell lysis. Although the temperate bacteriophage Acholeplasma virus L2 of Acholeplasma laidlawii is currently the only classified plasmavirus, related prophages reside in the genomes of different Acholeplasma species, where they are integrated into tRNA genes.
Table 1. Plasmaviridae. Characteristics of members of the family Plasmaviridae.
| Characteristic | Description |
| Typical member | Acholeplasma virus L2 (L13696), species Plasmavirus L2 |
| Virion | Enveloped, pseudo-spherical and pleomorphic virions (diameters 50–125 nm) |
| Genome | Circular, supercoiled dsDNA (11,965 bp) |
| Replication | DNA replication is bidirectional from two origins and is dependent on the host DNA replisome; virions are released by budding |
| Translation | Translational coupling or reinitiation may be involved in translation of the viral polycistronic mRNAs by the host translation machinery |
| Host range | Acholeplasma species |
| Taxonomy | Single genus with a single species |
Virion
Morphology
Virions are quasi-spherical, slightly pleomorphic, enveloped and about 80 nm (range 50–125 nm) in diameter (Figure 1. Plasmaviridae). At least three distinct virion forms are produced during infection (~75% of virions are 70–80 nm, ~20% are 80–90 nm, and ~5% are 110–120 nm). The three virion forms can be separated by velocity sedimenation and gel electrophoresis. The three forms have the same protein composition, albeit with differences in the relative amounts of each protein, but vary with respect to the number of encapsidated genome copies (from 1 to 3) (Poddar et al., 1985). Thin-sections show virions with electron-dense cores, presumably containing condensed DNA (Gourlay et al., 1973). Virion assembly is coupled to virion release from the infected cells (Maniloff and Dybvig 2006). The absence of a regular capsid structure suggests plasmavirus virions consist of a condensed nucleoprotein bounded by a proteinaceous lipid vesicle.
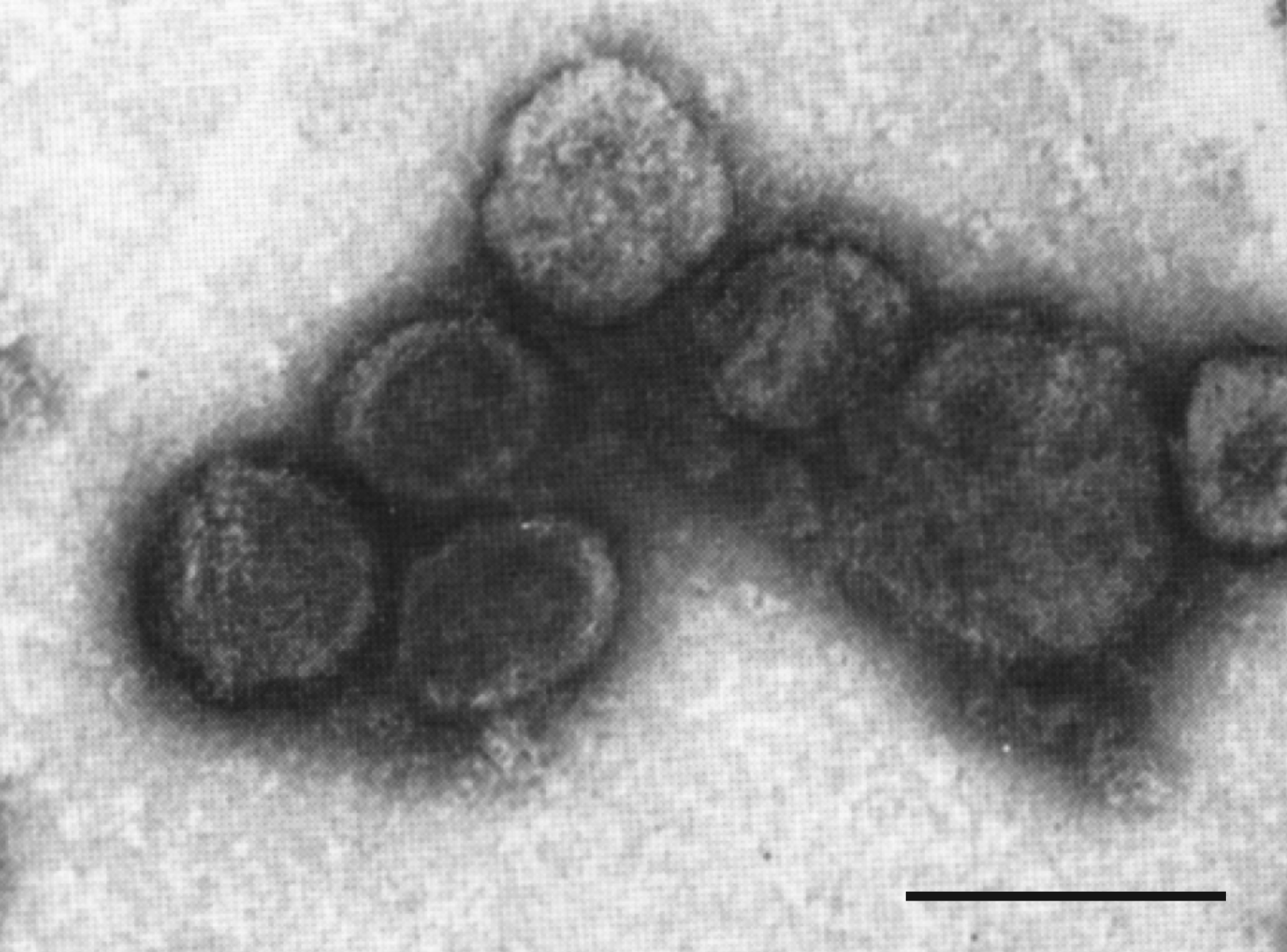 |
| Figure 1. Plasmaviridae. Electron micrograph of Acholeplasma virus L2 virions negatively-stained with uranyl acetate. Bar, 100 nm. Modified from (Gourlay 1971) with permission from Microbiology Society. |
Physicochemical and physical properties
Virions are extremely heat-sensitive, relatively cold-stable and inactivated by nonionic detergents (Brij-58, Triton X-100 and Nonidet P-40), ether and chloroform (Gourlay 1971, Gourlay 1973, Greenberg and Rottem 1979). Viral infectivity is resistant to DNase I and phospholipase A, but sensitive to pronase and trypsin treatment (Greenberg and Rottem 1979, Sladek and Maniloff 1983). UV-irradiated virions can be reactivated in host cells by excision and SOS DNA repair systems (Das et al., 1977). Virions are relatively resistant to photodynamic inactivation (Das et al., 1980).
Nucleic acid
The genome of Acholeplasma virus L2 (AVL2) consists of a circular, negatively supercoiled dsDNA molecule(s) of 11,965 bp, with a G+C content of 32% which is similar to that of its host (31.8%) (Maniloff et al., 1994). The genome is infectious when introduced into the cell interior (Sladek and Maniloff 1983).
Lipids
Lipid composition of the virions and host cell membranes are qualitatively similar, while fatty acid compositions are essentially identical (Greenberg and Rottem 1979, Putzrath et al., 1980). Variation of host cell membrane fatty acid composition leads to virions with corresponding fatty acid composition variations (Putzrath et al., 1980). Viral membrane lipids are in a bilayer structure (Greenberg and Rottem 1979, Putzrath et al., 1980).
Proteins
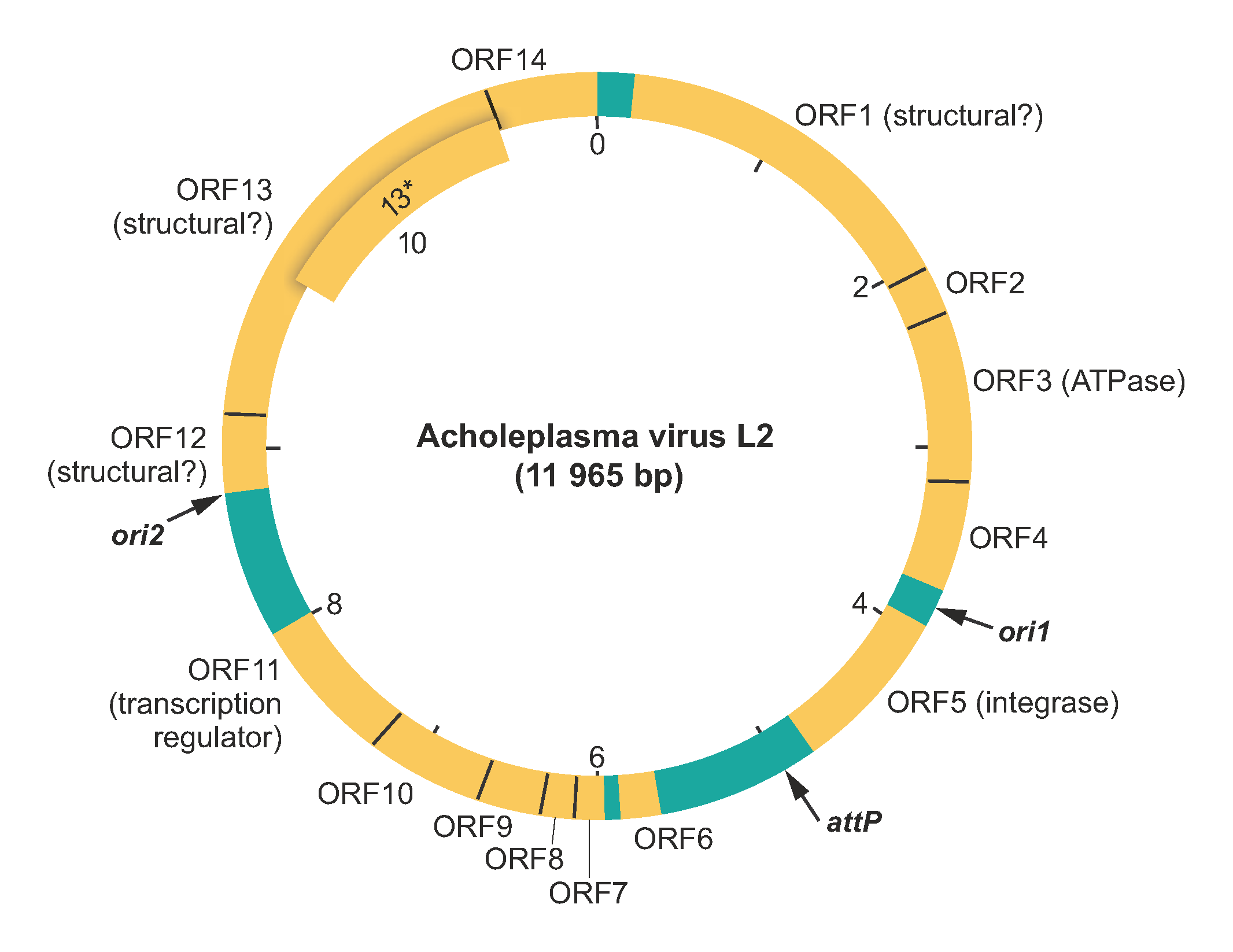
Virions contain at least four major proteins of about 64, 61, 58 and 19 kDa. Several minor protein bands are also observed in virion preparations (Poddar et al., 1985, Greenberg and Rottem 1979, Putzrath et al., 1980). The 64- and 61-kDa proteins are located on the outer surface of the virion and might be involved in the early stages of infection, including host recognition and binding (Greenberg and Rottem 1979, Rottem and Greenberg 1982)}. The 19 kDa protein has been suggested to be a DNA-binding protein involved in the assembly of the virion nucleoprotein core (Poddar et al., 1985, Putzrath et al., 1980). There is evidence that the 64-, 61- and 19-kDa proteins are encoded by open reading frame (ORF) 13, ORF1 and ORF12, respectively (Maniloff and Dybvig 2006) (Table 2). Consistently, sequence analysis has shown that the ORF13 protein contains a carbohydrate/sugar-binding domain, whereas ORF12 encodes a basic protein with a pI of 9.7. Most of the proteins are refractory to functional annotation using currently available sequence analysis methods. However, profile-profile comparisons suggest that ORF3 encodes an AAA+ ATPase, ORF5 encodes an integrase of the tyrosine recombinase superfamily, and ORF11 encodes a MarR-like transcriptional regulator containing a winged helix-turn-helix DNA-binding domain (Table 2).
Carbohydrates
None of the virion proteins gave a positive signal in periodic acid-Schiff reaction, suggesting that viral proteins are not glycosylated (Greenberg and Rottem 1979).
Genome organization and replication
The AVL2 genome contains 15 ORFs, all of which are encoded on the same strand and start with an ATG codon (Figure 2. Plasmaviridae, Table 2. Plasmaviridae). Each of the ORFs has an upstream Shine-Dalgarno sequence (Maniloff et al., 1994). At least 11 genes are translated from overlapping reading frames and are transcribed from at least eight promoters.
Table 2.Plasmaviridae. Genome annotation of Acholeplasma virus L2.
| ORF | Accession # | Mw (kDa) | TMD | Comments |
|---|---|---|---|---|
| ORF 1 | AAA87957 | 66,643 | 4 | putative viral envelope protein (61 kDa) |
| ORF 2 | AAA87958 | 9,620 | 2 | – |
| ORF 3 | AAA87959 | 37,157 | 1 | putative AAA+ ATPase |
| ORF 4 | AAA87960 | 18,224 | 1 | contains a domain related to the N-terminal domain of transcription termination factor Rho |
| ORF 5 | AAA87961 | 34,868 | putative integrase (tyrosine recombinase superfamily), gene is upstream of the attP site | |
| ORF 6 | AAA87962 | 9,799 | – | |
| ORF 7 | AAA87963 | 14,047 | – | |
| ORF 8 | AAA87964 | 7,412 | – | |
| ORF 9 | AAA87965 | 9,332 | – | |
| ORF 10 | AAA87966 | 16,143 | – | |
| ORF 11 | AAA87967 | 25,562 | MarR-like transcriptional regulator, winged helix-turn-helix domain | |
| ORF 12 | AAA87968 | 17,214 | 4 | basic protein, putative major virion DNA-binding protein (19 kDa) |
| ORF 13 | AAA87969 | 81,308 | 1 | putative integral membrane protein, has 27 amino acid N-terminal peptidase cleavage signal sequence; carbohydrate/sugar-binding domain (78 kDa) |
| ORF 13* | 47,699 | 1 | translation start site is 295 codons downstream from ORF13 start site and in same reading frame | |
| ORF 14 | AAA87956 | 26,105 | 2 | has 26 aa N-terminal peptidase cleavage signal sequence; putative envelope protein |
(Abbreviations: ORF, open reading frame; TMD, transmembrane domain; Mw, molecular weight)
AVL2 DNA replicates bidirectionally from two ori sites, each containing a DnaA box bounded by AT-rich 6-mer repeats (Poddar and Maniloff 1987). Both ori sites are located within intergenic regions of the virus genome. Replication of the parental DNA is membrane-associated and depends on the host DNA replisome, including DNA polymerase III and DNA gyrase (Poddar and Maniloff 1984, Maniloff 1988). Some parental DNA is found in progeny virions, suggestive of semiconservative replication of infecting parental DNA.
AVL2 infection involves a non-cytocidal productive infectious cycle (rise period) followed by a lysogenic cycle in each infected cell. Virus DNA replication in infected cells continues throughout the viral rise period, but not all intracellular progeny DNA is packaged into virions. AVL2 DNA replication and progeny virion maturation continue for several hours after integration of a phage genome into the host cell genome. Although progeny DNA replication stops about 5-6 hours post infection, cytoplasmic progeny DNA persists up to at least 10 hours post-infection (Maniloff 1988, Dybvig and Maniloff 1983).
Biology
AVL2 adsorption presumably involves virus-cell surface interaction followed by the fusion of the viral and host membranes, although the nature of viral and cellular components involved in either process remain obscure (Maniloff 1988). However, the lipoglycan oligosaccharide may form part of the receptor for AVL2 adsorption (Al-Shammari and Smith 1982). Fusion of the viral and host membranes presumably results in delivery of the nucleoprotein core into the cell interior (Maniloff and Dybvig 2006).
In non-cytocidal infection, progeny viruses are released by budding from the host cell membrane, with the host surviving as a lysogen. Lysogeny involves site-specific integration of the AVL2 genome into a unique site in the host cell chromosome (Dybvig and Maniloff 1983). The putative AVL2 attachment site (attP; CATCTTCAT–7nt–CTGAAGATA) is located in the intergenic region downstream of the integrase gene. Lysogens are resistant to superinfection by homologous virus but not by heterologous virus, and are inducible by UV-irradiation and mitomycin C treatment (Putzrath and Maniloff 1978). Although newly infected cells grow slower than uninfected cells, eventually lysogens have the same growth rate as uninfected cells (Maniloff 1988). Progeny virions appear to be released by budding through the cell membrane without causing cell lysis (Gourlay et al., 1973, Maniloff et al., 1977).
AVL2 infects Acholeplasma laidlawii strains. Other putative plasmaviruses have been reported to infect A. laidlawii (Acholeplasma phages v1, v2, v4, v5 and v7), A. modicum (Acholeplasma phage M1) and A. oculi strains (Acholeplasma phage O1), but their genome sequences are not available. Analysis of the available Acholeplasma genome sequences showed that apparently complete prophages related to AVL2 are present in several strains of A. oculi, A. brassicae, A. hippikon, A. palmae and A. axanthum. Similar to AVL2, these proviruses encode integrases and are integrated into various tRNA genes.
Derivation of names
Plasma: from the Greek plasma, “shaped product”, referring to the plastic virion shape.
Relationships within the family
No information available.
Relationships with other taxa
Except for the integrase, plasmaviruses do not share homologous proteins with other known viruses. Although virion organization and morphology of plasmaviruses appear to be superficially similar to that of archaeal viruses of the family Pleolipoviridae (Bamford et al., 2017), this resemblance is likely to be a result of convergent evolution rather than shared ancestry (Krupovic et al., 2018). Furthermore, similar virion organization has been reported for an unclassified mycoplasma bacteriophage L172 (Dybvig et al., 1985). However, the genome of the latter virus consists of a single-stranded DNA and its sequence is not available, precluding further assessment of the evolutionary relationship.