Family: Matonaviridae
Annette Mankertz, Min-Hsin Chen, Tony L. Goldberg, Judith M. Hübschen, Florian Pfaff, Rainer G Ulrich
The citation for this ICTV Report chapter is the summary that has been published as Mankertz et al., (2022): ICTV Virus Taxonomy Profile: Matonaviridae 2022, Journal of General Virology (2022) 103:001817
Corresponding author: Annette Mankertz ([email protected])
Edited by: Peter Simmonds and Stuart G. Siddell
Posted: November 2022
Summary
Matonaviridae is a family of small, enveloped viruses with single-stranded positive-sense RNA genomes of approximately 9.6–10 kb (Table 1.Matonaviridae). The single genus Rubivirus includes rubella virus (RuV, species Rubivirus rubellae) infecting humans, ruhugu virus (RuhV, species Rubivirus ruteetense) infecting bats and rustrela virus (RusV, species Rubivirus strelense) infecting wild rodents and other mammals. The structural and functional description of matonaviruses is mainly based on the characterization of RuV, since virus isolates are not yet available for RuhV and RusV.
RuV is transmitted via the respiratory route. Postnatal infection typically leads to a relatively benign disease in children and adults, characterized by rash and fever. In contrast to postnatal infection, which is only rarely associated with complications, RuV is an important teratogen: infection of seronegative women during the first trimester of pregnancy confers a high risk of miscarriage, stillbirth, or severe foetal malformations, known as congenital rubella syndrome (CRS). There is also an association between persistent RuV infection and granulomatous disease in adults. Safe and effective live-attenuated vaccines against rubella are globally available; RuV has been targeted for elimination by the WHO.
The zoonotic potential of RuhV and RusV is not yet known. Guangdong Chinese water snake rubivirus, tiger flathead matonavirus and tetronarce matonavirus are unclassified viruses that have been reported to be related to members of the family Matonaviridae.
Table 1. Matonaviridae. Characteristics of members of the family Matonaviridae.
|
Characteristic |
Description |
|
Example |
rubella virus (JN635296), species Rubivirus rubellae, genus Rubivirus |
|
Virion |
Enveloped, 50–90 nm pleomorphic virions, spherical to tube-like, with a single capsid protein and two envelope glycoproteins |
|
Genome |
9.6–10 kilobases of non-segmented, positive-sense RNA |
|
Replication |
Cytoplasmic, in vesicles derived from the plasma membrane/endosomal compartment. Assembled virions bud into the lumen of the Golgi apparatus |
|
Translation |
Non-structural proteins are translated from the genomic RNA, and structural proteins from a subgenomic RNA, both as polyprotein precursors |
|
Host range |
Humans (rubella virus, bats (ruhugu virus), mammals living in a zoo and rodents (rustrela virus); unclassified matonavirus-like viruses have been detected in fish and reptile species |
|
Taxonomy |
Realm Riboviria, kingdom Orthornavirae, phylum Kitrinovircota, class Alsuviricetes, order Hepelivirales: one genus (Rubivirus) including three species Rubivirus rubellae, Rubivirus ruteetense and Rubivirus strelense. |
Virion
Morphology
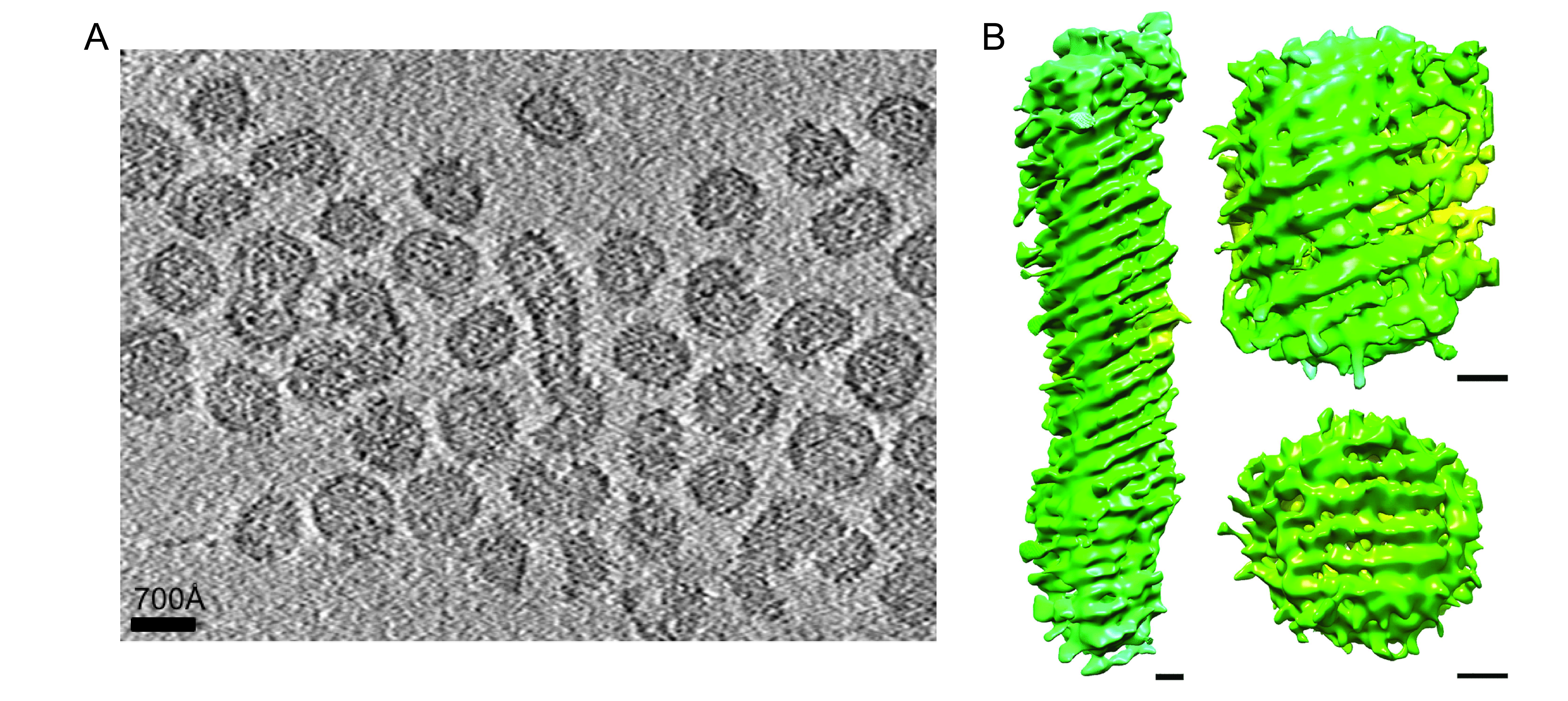
RuV particles are pleomorphic, ranging from spherical to tube-like in shape. Particle sizes vary between 50 and 90 nm in length and width (Battisti et al., 2012) with an electron-dense nucleocapsid core, a lipid bilayer, and surface glycoproteins (Figure 1.Matonaviridae). The virions have an outer shell, about 90–130 Å in width, that is composed of the cell-derived membranes and includes the viral glycoproteins. The nucleocapsid consists of one copy of the viral genome to which capsid protein monomers are bound. The outer shell and nucleocapsid are separated on average by about 70 Å (Battisti et al., 2012, Mangala Prasad et al., 2013). Data on RuhV and RusV particles are not yet available.
The glycoproteins, E1 and E2, form heterodimers that are arranged in rows on the particle surface. There are 4–6 rows of glycoproteins and some wrap around the particle forming a helical arrangement. This is unique among enveloped viruses, as similar helical arrangements have previously only been observed for matrix proteins or nucleoprotein complexes, not surface glycoproteins. There is a gap between the lipid bilayer and the nucleocapsid core, as seen with flaviviruses. The internal core does not show icosahedral symmetry unlike alphavirus particles. Rather the capsid (C) protein is arranged as a homodimer in a grid-like pattern, often underneath the glycoprotein spikes (Battisti et al., 2012, Mangala Prasad et al., 2017).
 |
|
Figure 1.Matonaviridae. Images and structures of RuV particles. (A) Purified RuV flash frozen in vitreous ice (courtesy of AJ Battisti and V. Mangala Prasad). (B) Representation of three different rubella virions, each determined using cryo-electron tomography without averaging procedures. The virions are different in size and shape but all show surface glycoproteins arranged in rows around the particle. Bars: 100 Å. The resolution of the reconstructions are not absolute but are estimated to be better than 50 Å (reproduced and modified from (Mangala Prasad et al., 2017). |
Physicochemical and physical properties
The buoyant density of RuV in sucrose gradients is 1.18–1.19 g cm-3. Reported sedimentation coefficients for RuV range from 240S to 350S. This wide range is due to contamination with host proteins and lipids associated with virions during purification. RuV can be physically denatured by heat inactivation and exposure to ultraviolet light and treatment with detergents and various chemicals including acids, urea, formaldehyde, and beta-propiolactone. RuV particles are stable at physiological pH ranges and can be stored at –80°C without significant loss of infectivity (Parkman 1965, Hobman 2013, Kuhn 2013).
Nucleic acid
Rubiviruses (i.e. members of the genus Rubivirus) have non-segmented single-stranded, positive-sense RNA genomes. The genome of rubella virus strain RVi/Bismarck.ND.USA/23.08/2B (JN635296) is 9761 nucleotides in length, although there can be one or two nucleotide insertions or deletions in the intergenic region (IR). In-frame deletions in protein coding regions of granuloma-associated RuV have also been reported (Perelygina et al., 2019b). The genomes of RuhV and RusV are comprised of 9621 and 9631 nucleotides, respectively (Bennett et al., 2020a, Bennett et al., 2020b, Pfaff et al., 2022). The genomic RNA of RuV contains 30 mol% guanine residues and 39 mol% cytosine residues; the G+C content of RusV reaches 85 mol% in the intergenic region and the N-terminus of the C protein-encoding region, this being the highest G+C content of all known RNA viruses.
Proteins
RuV consists of three structural proteins, the C protein, that forms a complex with the genome, and the two glycoproteins E1 and E2 that are exposed on the virus envelope. E1, a class II membrane fusion protein, is the main antigen and contains neutralizing epitopes. E2 is responsible for viral attachment to the cellular receptor. E1 and E2 form initially a homodimer. Upon acidification of the endosome during uptake, the concomitant trimerization of the E1 subunits is induced. This E1 homotrimer is fusion active and promotes release of the viral nucleocapsid in cytoplasm after endosome and viral membrane fusion.
Lipids
RuV acquires its lipid membrane from the host cell during budding. In general, the lipid membrane accounts for close to 30% of the virion mass.
RuV assembly and budding occurs at the Golgi complex (Risco et al., 2003) and the plasma membrane depending on the host cell. Newly budded, intracellular particles in BHK21 cells were electron-dense and considered “immature” (Bardeletti et al., 1979). Virion maturation occurs within the Golgi (Bardeletti et al., 1979), where the nucleocapsid core condenses and undergoes structural reorganizations. The mature particles exhibit a double shell-like structure (Mangala Prasad et al., 2017) and are secreted to the extracellular environment.
Carbohydrates
The RuV proteins E2 and E1 are both glycosylated. E1 has three N-linked residues and E2 has three N-linked and O-linked carbohydrate moieties (Oker-Blom et al., 1984, Lundstrom et al., 1991). N-linked glycosylation has a role in E2 glycoprotein processing, disulphide bond formation, stability, and intracellular transport to the Golgi compartment for budding (Qiu et al., 1992). Two isoforms of E2 have been reported to be translated, each with differing levels of glycosylation (Frey 1994).
Genome organization and replication
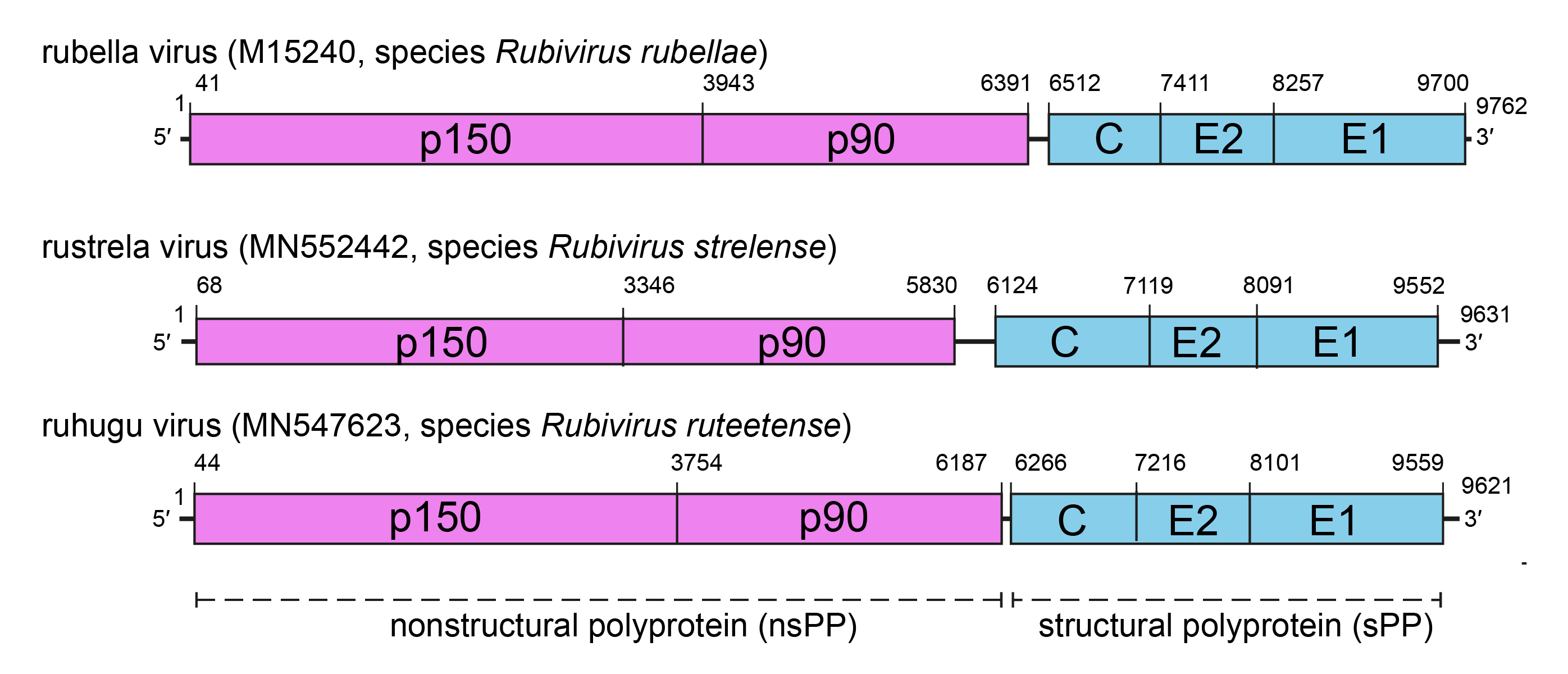
Rubivirus genomes have two non-overlapping polycistronic open reading frames (ORFs) with non-coding regions (NCRs) at the 5′- and 3′-termini, and an intergenic region between the two ORFs (Figure 2.Matonaviridae). The UTRs and the IR are thought to form secondary structures that can be bound by proteins that initiate and regulate transcription and replication of RuV. The 5′-proximal ORF encodes the two non-structural proteins p150 and p90, responsible for virus RNA transcription and replication. The 3′-ORF encodes three structural proteins, namely C protein, E1 and E2. Both ORFs are expressed as polyprotein precursors (PP).
RuV replication is comparatively slow. Even at a high multiplicity of infection, cells are not uniformly infected after 24 h. This hints at a cell cycle-associated factor that might regulate events in RuV entry or early replication. RuV genome replication initiates with the synthesis of negative-sense RNA that is complementary to the genomic sequence. This RNA serves as a template for both genomic and subgenomic RNA synthesis. Both positive-sense RNAs, genomic and subgenomic, serve as mRNAs for translation of the non-structural polyprotein precursor (nsPP) and the structural polyprotein precursor (sPP), respectively. The subgenomic RNA contains approximately 1/3 of genomic sequence from the 3′-end and is transcribed from the genome length negative-sense RNA during replication of RuV. RNA synthesis is driven by the subgenomic RNA promoter present in the IR (Tzeng and Frey 2002). Both the genomic and subgenomic RNAs have a viral type 0 7meGpppA cap at their 5ʹ-termini (Ahola and Kaariainen 1995), and a poly-A tail at their 3ʹ-termini. Only the genomic RNA is packaged into the viral particles and the encapsidation signal for binding of C protein to the genomic RNA is mapped to nucleotides 347–375 (Liu et al., 1996). The negative-sense RNA of the replication intermediate is neither capped nor contains a poly-A tail (Kuhn 2013).
Viral replication occurs in association with cellular membranes leading to formation of viral replication factory complexes. These compartments are surrounded by rough endoplasmic reticulum (ER) and can be stained with antibodies against p150, dsRNA, and lysosomal markers. Mitochondria are relocated to cluster in the vicinity of the replication complexes.
Proteins
RuV virions contain three structural proteins, C protein and two glycoproteins E1 and E2. Transcription and replication are performed by the two non-structural proteins p150 and p90. E1 and E2 are responsible for receptor binding, membrane fusion and cell entry (Hobman 2013, Kuhn 2013, Das and Kielian 2021). The genome of RuV is complexed with C protein and thereby protected from damage or degradation. The amino acid (aa) sequences of RuhV and RusV C proteins show 46% and 51% similarity to that of RuV, respectively. The E1 glycoproteins of RuhV and RusV display 56% and 51% amino acid similarity to RuV, while the values for E2 are lower at 43% and 31%. The RNA-directed RNA polymerase (RdRP) amino acid sequences are more conserved showing 75.7% and 65.4% similarity for RuhV and RusV in comparison to RuV.
The 5′-proximal ORF of RuV encodes a nsPP of 2116 aa. This precursor is cleaved into the two non-structural proteins, p150 (150 kDa, 1301 aa) and p90 (90 kDa, 905 aa). The cleavage of the polyprotein happens autocatalytically by a virus-encoded papain-like cysteine protease domain encoded in the carboxyl portion of p150. Both non-structural proteins are required for genome replication: p150 contains a methyltransferase domain, a Y domain (of unknown function), an X domain (ADP-ribose-1″-phosphatase), and a protease. p90 fulfils two molecular functions as helicase and RdRP, with the tripeptide GDD at the active centre (aa residues 1965–1967) of the replicase. Interestingly, the arrangement of the respective domains is different from that of the alphaviruses and similar to that of hepeviruses. The subcellular localization of p90 was described as cytoplasmic punctiform structures forming linear chains, while p150 was associated with tube-like structures which are involved in synthesis of viral RNA (Kujala et al., 1999).
The 3′-proximal ORF encodes a sPP which is translated from a 24S sub-genomic RNA that is transcribed after infection of the host cell. The 1063 aa precursor is synthesized at the ER membrane, cleaved by the host cell signal peptidase into C protein, E2, and E1 and translocated to intracellular membranes (Kuhn 2013). C protein is a multifunctional protein of 300 aa (33–35 kDa) with a high fraction of arginine and proline residues, resulting in a net positively charged protein. Upon generation of a sufficient amount of C protein, it assembles with the negatively charged viral RNA to form the viral nucleocapsids. Moreover, C protein forms disulphide-linked homodimers and interacts with the cytoplasmic tails of E1 and E2. C protein has been reported to perform multiple functions in viral assembly, but also in viral transcription and replication. Phosphorylation of serine 46 in the RNA binding domain in C protein reduces RNA binding and thus regulates virus assembly (Law et al., 2006). The expression of C protein, E1 and E2 in absence of the viral genome results in formation of virus-like particles (VLPs), indicating that nucleocapsid formation and budding of virus particles are not tightly linked. Moreover, C protein interacts with two mitochondrial proteins, p32 and Par-4 (Beatch and Hobman 2000).
E1 comprises 481 aa and has a mass of 59 kDa, whereas the mass of E2 (282 aa) ranges from 44 to 50 kDa due to differential glycosylation. In contrast to alphaviruses, where E2 does not retain the membrane anchor and is located in the cytoplasm, the E2 signal peptide of RuV remains attached to the C-terminus of RuV C protein and serves as a membrane anchor. In front of this element is a highly structured domain named “C-terminal domain” (CTD) which is also present in RuhV and RusV E2. Both E1 and E2 are palmitoylated and both are N-linked and O-linked type I glycosylated membrane proteins (Lundstrom et al., 1991, Qiu et al., 1992, Hobman 2013). They dimerize to form the functional spike complexes found on the virion surfaces. The entry of RuV is mediated by receptor binding and endocytosis. E2 and E1 are involved in host cell binding and membrane fusion. Myelin oligodendrocyte glycoprotein (MOG) serves as a receptor for RuV by binding to E1 (Cong et al., 2011), but MOG-independent RuV infection of HaCaT keratinocytes has also been described (Trinh et al., 2018). E1 is a class II fusion protein that requires endosomal levels of calcium ions for fusion (DuBois et al., 2013, Dube et al., 2014, Dube et al., 2016), unlike alphavirus and flavivirus class II fusion proteins (Vaney and Rey 2011). The residues involved in the calcium-coordinated membrane fusion are conserved among RuV and RuhV (Bennett et al., 2020b). E1 contains the dominant antigenic determinant and the target of neutralizing antibodies.
 |
|
Figure 2.Matonaviridae Genomic organization of three representatives of the family Matonaviridae. Comparative genome architecture of RuV, RuhV and RusV, showing the ORFs encoding the non-structural polyprotein (nsPP, pink), the structural polyprotein (sPP, blue) and the boundaries of processed products. |
The structural proteins for RuV are inserted into the ER during translation and are translocated to intracellular membranes (Kuhn 2013). C protein assembles with the viral RNA. The assembly occurs during the budding process associated with the Golgi apparatus. Budding through the Golgi and plasma membrane leads to the acquisition of a lipid envelope containing the two main viral membrane glycoproteins (Frey 1994).
Biology
Host range
RuV is known to infect only humans; no animal reservoir has been described. Moreover, animal models for RuV are not currently available. RuhV was found in apparently healthy bats (Cyclops leaf-nosed bat, Hipposideros cyclops) in Uganda. RusV was found in apparently healthy rodents (yellow-necked field mouse, Apodemus flavicollis) in Germany at high prevalence, suggesting these animals to be the reservoir hosts. Moreover, RusV causes severe encephalitis and was found in brain tissues of four acutely ill zoo animals: a donkey (Equus asinus), a capybara (Hydrochoerus hydrochaeris), a red-necked wallaby (Macropus rufogriseus), a South American coati (Nasua nasua), and a wild Eurasian or European otter (Lutra lutra) (Bennett et al., 2020b, Pfaff et al., 2022). RusV was subsequently detected in brain tissues of yellow-necked field mice in the zoo and its surroundings, suggesting spillover from mice to these other species. There are sequences of related but not yet classified matonavirus-like viruses detected in fish and reptile species: Guangdong Chinese water snake rubivirus (Shi et al., 2018b), tiger flathead matonavirus (Geoghegan et al., 2021) and tetronarce matonavirus from the pacific electric ray (Grimwood et al., 2021) (see “Related, unclassified viruses” section).
Transmission
RuV is transmitted from person-to-person, primarily through respiratory routes (Kuhn 2013). The zoonotic potential of RuhV and RusV is currently unclear. Their route of transmission is also not yet clear.
Geographical distribution
RuV has a global distribution but it is much less prevalent in countries where vaccination programs are in place. RuhV has to date been found only in Uganda and RusV only in Germany, although the geographic range of these viruses may be wider and follow the distributions of their likely reservoir hosts. Guangdong Chinese water snake rubivirus was detected in South China (Shi et al., 2018b), tiger flathead matonavirus in Australia (Geoghegan et al., 2021) and tetronarce matonavirus from the pacific electric ray in the USA (Grimwood et al., 2021), but are yet unclassified.
Pathogenicity
Postnatal rubella, formerly known as German measles, is generally mild and 30–50% of cases are subclinical. Symptomatic rubella can involve maculopapular rash, lymphadenopathy, low-grade fever, conjunctivitis, sore throat, and arthralgia. Rare complications are thrombocytopenia or encephalitis. RuV is a potent teratogen: children of unvaccinated women infected with RuV during the first trimester of pregnancy are at a 90% risk for being born with congenital rubella syndrome (CRS). Birth defects due to CRS include deafness, cataracts, and heart defects (Gregg 1947). CRS infants shed high amounts of virus for up to twelve months. In addition, persistent RuV infection has been reported to be associated with diseases such as Fuchs' uveitis, fatal encephalitis, and granuloma (Lazar et al., 2016, Perelygina et al., 2019a).
Attenuated live vaccines against rubella were developed in the 1960s. Since then, they have been used to vaccinate millions of children and adults. Since there is no animal reservoir, it may be possible to eradicate RuV. Consequently, rubella and CRS have been targeted for elimination by the WHO (WHO 2012). Transmission has been interrupted in several countries or even continents for prolonged periods, e.g. rubella was declared by PAHO/WHO to be eliminated in the Americas in 2015 (PAHO 2015). Interestingly, vaccination against rubella in early pregnancy does not lead to CRS, indicating the attenuation process has led to the loss of the teratogenic potential of the vaccine strains.
RuhV was detected in oral swabs of healthy bats and has not been associated with disease to date. RusV was detected in the brain tissues of apparently healthy wild yellow-necked field mice and in acutely encephalitic necropsied animals. Histopathology examination revealed a non-suppurative meningoencephalitis, characterized by perivascular cuffing, meningeal infiltrates, and glial nodules. Thus, the pathogenicity of RusV appears to vary widely between the reservoir host (yellow-necked field mice) and mammals of other species infected by spill-over events.
RuV can be propagated in vitro, on e.g. Vero, RK13 or BHK21 cells. In tissue culture, RuV replicates in the cytoplasm of infected cells. RuV infection does not appear to inhibit cell macromolecular synthesis. Microscopically, cells infected with wildtype RuV appear similar to uninfected cells and produce no significant cytopathic effect; however, rearrangements of cellular cytoskeletal elements and organelles such as mitochondria have been reported. Moreover, RuV infection induced a shift of the bioenergetic state of epithelial cells (Vero and A549) and human umbilical vein endothelial cells to a higher oxidative and glycolytic level (Bilz et al., 2018).
Antigenicity
Only one serotype of RuV has been identified. E1 is immunodominant although antibodies to other viral proteins can also be detected in vaccinated individuals (Haralambieva et al., 2017). Amino acid sequences in neutralization and hemagglutination epitopes in E1 show no significant differences among currently circulating strains. RuV vaccines provide sufficient protection against wildtype clade 1 and 2 viruses, members of these two clades differing by 8–10% in the nucleotide sequence of a 731 region of the E1 gene. Antigens derived from a single RuV strain can be used to detect antibodies induced by all strains in serological tests used for screening and diagnosis (Best et al., 1992). Data on the antigenicity of RuhV and RusV are not yet available, although tertiary structure models of trimeric E1 of both viruses show structural homology to RuV. Amino acid sequences of RuV, RuhV, and RusV are moderately to highly conserved within 4 putative B-cell epitopes in the E1 protein and, in the case of RuV and RuhV viruses, within two putative T-cell epitopes in the capsid protein (Perelygina et al., 2019b).
Derivation of names
Matonaviridae: after George de Maton who in 1814 first distinguished rubella from measles and scarlet fever.
Rubivirus: from Latin rubeus meaning “reddish”.
Rubivirus rubellae; from rubella virus: from Latin “little red”
Rubivirus ruteetense; from ruhugu virus, named for its rubivirus-like genome and the Tooro word for insectivorous bat, obuhuguhugu, and Ruteete Subcounty, Uganda, where ruhugu virus was first detected.
Rubivirus strelense; from rustrela virus, itself named for its rubivirus-like genome and from the German Strela Sound of the Baltic Sea, where the virus was first detected.
Relationships within the family
The RuV genome is very stable. More than 80 genomes of RuV have been sequenced and all are nearly identical in terms of genome length and the lengths of the coding and noncoding regions. Short deletions are occasionally encountered in the intergenic region. Based upon the nucleotide sequence of a 731 region of the E1 gene, RuV genomes are grouped according to the WHO proposed nomenclature system into two clades and several genotypes (1A-J and 2A-C); these do not display different pathology or antigenicity, although their geographical distributions differ.
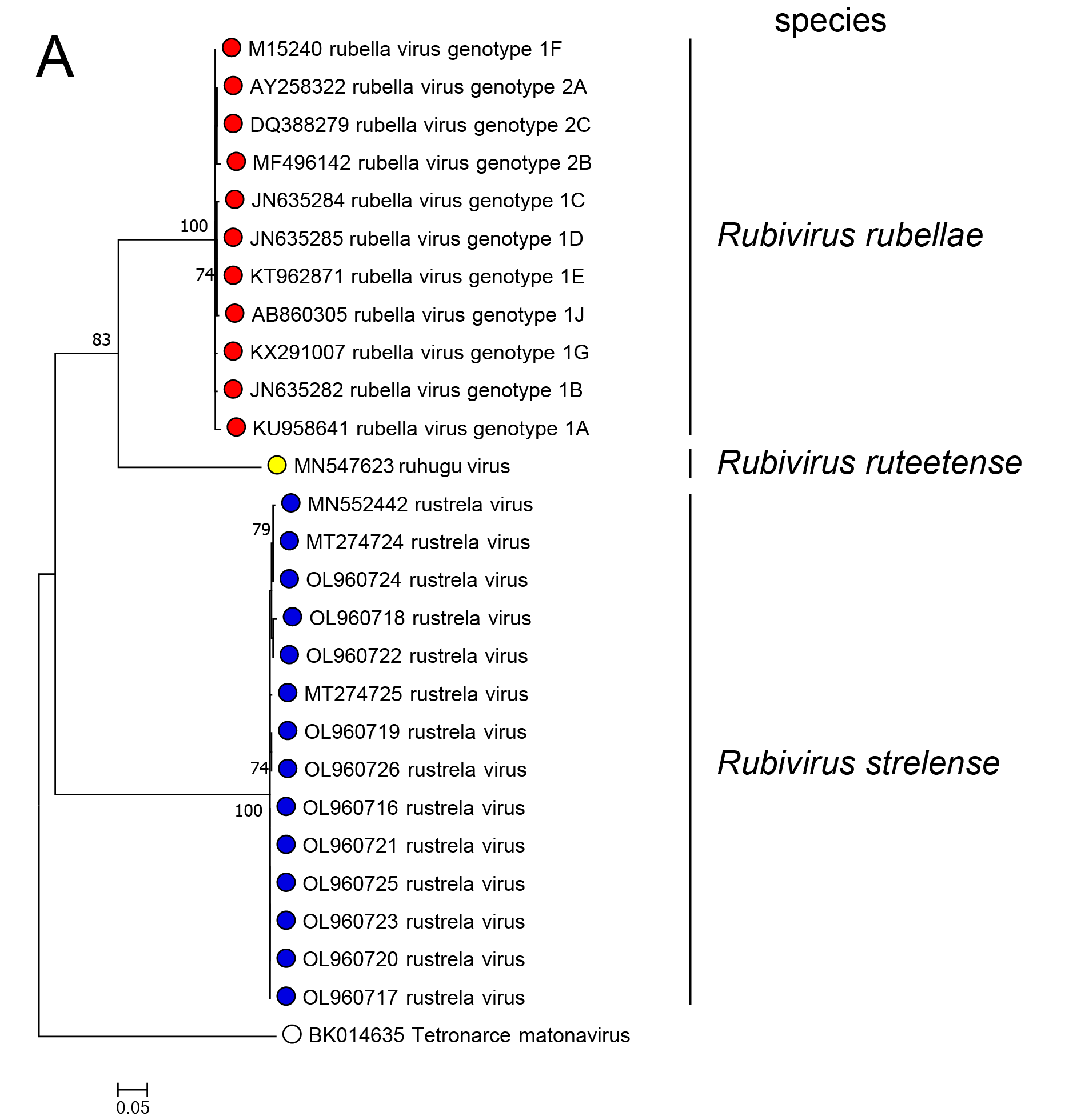
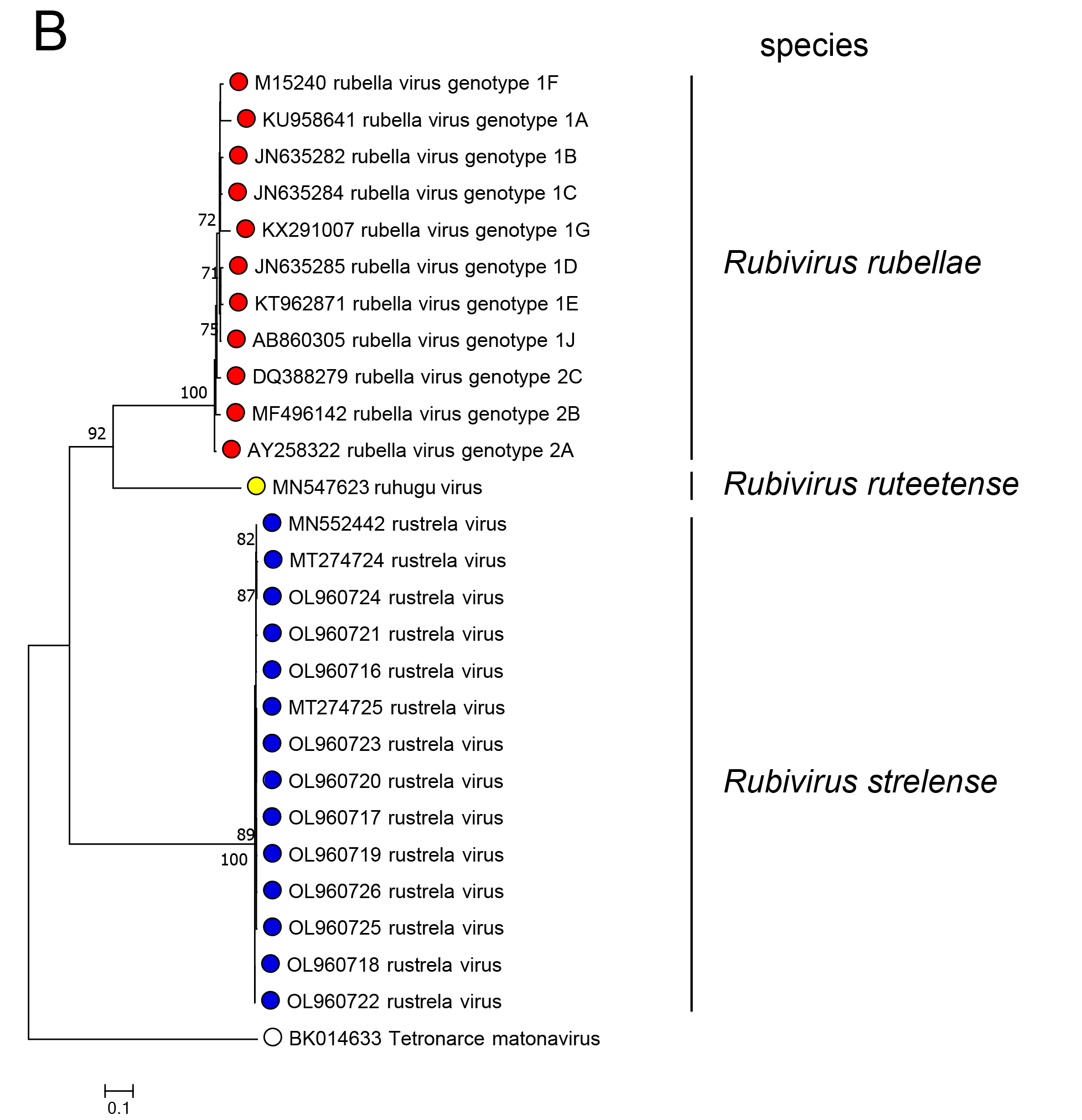
In phylogenetic analysis of part of the RdRP or of the C-E2-E1 polyprotein, RuhV forms a sister group to RuV and both share a common ancestor (Figure 3.Matonaviridae). RusV forms a more distant group that shares a common ancestor with RuV and RuhV. This is consistent with the greater divergence of RusV from RuV for each of the five mature virus proteins. Related, unclassified viruses have been detected in fish and reptile species (see below).
 |
 |
|
Figure 3.Matonaviridae Phylogenetic relations of viruses in the family Matonaviridae. (A) part of the RNA-directed RNA polymerase within the non-structural polyprotein (nsPP) p200 (399 aa, positions 1718 – 2116 in RuV) and (B) the complete p110 structural polyprotein (sPP, 1234 aa) with C (capsid) and envelope proteins E2 and E1, were aligned using MUSCLE (version 3.8.425) with 100 iterations, respectively. Maximum-likelihood phylogenetic trees were calculated with iqtree (version 2.3.1), using automated model selection and one million ultra-fast bootstrap replicates. The unclassified virus Tetronarce matonavirus was used as an outgroup. Numbers at nodes indicate bootstrap support values over 70%. The bar represents substitutions per site. |
Relationships with other taxa
The genus Rubivirus was previously classified in the family Togaviridae along with members of the genus Alphavirus because of similarities in virion properties. However, rubiviruses differ from alphaviruses in their mode of transmission, virion structure, and sequence relationships, and therefore have been moved to a separate family. Phylogenetic analysis of the RdRP of alphaviruses, RuV and those of other positive-sense RNA viruses demonstrates similarities with members of the families Benyviridae, Hepeviridae and Alphatetraviridae (Shi et al., 2018a). Members of the family Flaviviridae were also once classified with RuV in the family Togaviridae on the basis of biological and virological similarities but were moved to their own family because of differences in genetic organisation and mode of replication. However, similarities in the function and structural organization of the replication proteins among members of RNA viruses from several plant families implies that assignment to an alphavirus-like higher taxonomic rank may be appropriate (Rozanov et al., 1992).
Related, unclassified viruses
|
Virus name |
Accession |
Abbreviation |
|
Guangdong Chinese water snake rubivirus |
GCWV |
|
|
Tetronarce matonavirus |
TeMV |
|
|
tiger flathead matonavirus |
TiFV |
Virus names and virus abbreviations are not official ICTV designations.
These matona-like viruses have been detected in fish and reptile species through the use of metagenomic technologies. Guangdong Chinese water snake rubivirus was discovered via a large virological survey of vertebrates in China, tiger flathead matonavirus through a metatranscriptomic exploration of Australian marine fish (Geoghegan et al., 2021) while Tetronarce matonavirus was assembled in a transcriptome of a Pacific electric ray (Grimwood et al., 2021).

