Family: Artiviridae (Interim Report)
This is a summary page created by the ICTV Report Editors using information from associated Taxonomic Proposals and the Master Species List.
Edited by: Holly Hughes
Posted: October 2024
Summary
The family Artiviridae includes dsRNA viruses of invertebrates (Table 1 Artiviridae). The family Artiviridae was established in 2024 (Master Species List #39). The family currently has a single genus, Artivirus, with 9 species.
Table 1 Artiviridae. Characteristics of members of the family Artiviridae.
| Characteristic | Description |
| Example | Omono River virus (AB555544), species Artivirus shi, genus Artivirus |
| Virion | Non-enveloped, icosahedral particles of 40 nm diameter |
| Genome | One linear segment of dsRNA of 6.6–8.3 kbp (Figure 1 Artiviridae) |
| Replication | Unknown |
| Translation | Possibly involving −1 frameshifting |
| Host range | Invertebrates |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Duplornaviricota, class Chrymotiviricetes, order Ghabrivirales: 1 genus and 9 species (Figure 2 Artiviridae) |
Virion
Morphology
Artivirids have been described as icosahedral virions 40nm in diameter without any envelope (Poulos et al., 2006, Isawa et al., 2011). The 120-subunit capsid (T=1) has fiber-like surface protrusions extending 80 angstroms (Tang et al., 2008).
Nucleic Acid
The genome comprises a single segment of dsRNA, 6.6–8.3 kb in length, and encodes two open-reading frames. The presence of a pseudoknot conserved in artiviruses that results in a −1 ribosomal frame shift is suggested to produce a precursor protein (Zhai et al., 2010).
Proteins
Artiviruses have been demonstrated to produce a capsid protein about 99 kDa, an RNA-dependant RNA polymerase (RdRP) of 192 kDa, and three protein cleavage products that have similarity to dsRNA binding proteins (Poulos et al., 2006) and potential capsid-fiber complexes involved in cell entry or pathogenesis (Tang et al., 2008).
Genome organization and replication
The complete replication strategy of artivirids has not been elucidated, however several features of replication have been described. The fiber-like protrusion proteins are suggested to bind extracellular receptors and play a role in transmission (Shao et al., 2021). Endocytosis, and acidification are believed to cause dissociation of the protrusion protein and confirmational changes that facilitate replication (Shao et al., 2021). The untranslated regions of viral RNA have complementary nucleotides that are predicted to form panhandle-like structure, suggesting a binding site for RdRP (Zhai et al., 2010). Similar structures have been identified in dsRNA viruses in the order Reovirales. While two non-overlapping ORFs are present, the downstream ORF encoding the RdRP is believed be expressed as a fusion of the capsid and RdRP proteins produced from a −1 ribosomal frame shift (Zhai et al., 2010) (Figure 1 Artiviridae). The artivirid genome contains at least one 2A-like motif for ribosomal skipping and cleavage of viral polyprotein precursors (Isawa et al., 2011). This suggests co-translational cleavage of the polyprotein into the capsid, dsRNA binding protein, protrusion protein, and RdRP (Zhai et al., 2010).
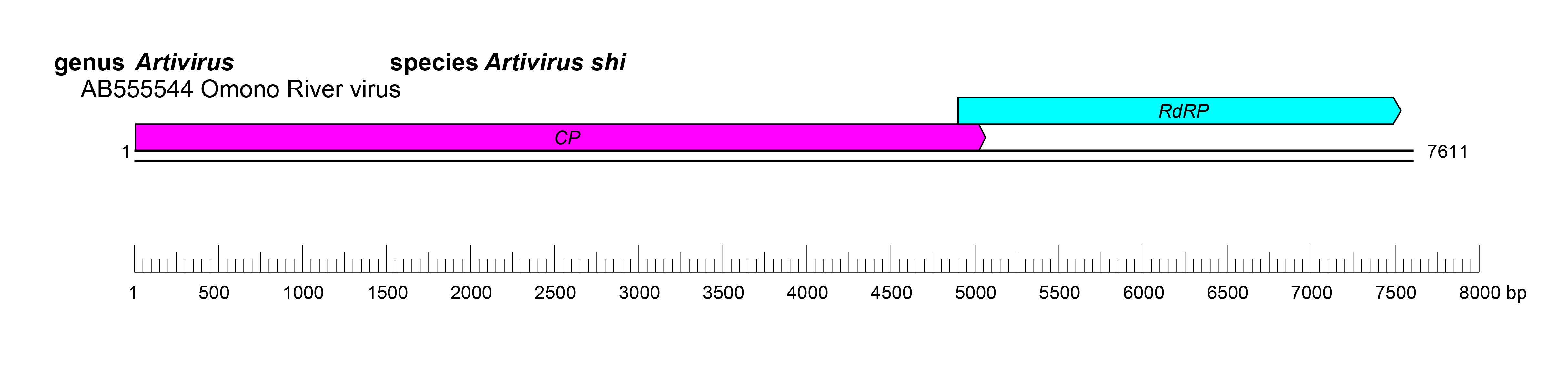 |
| Figure 1 Artiviridae. Genome organisation of Omono River virus, a member of the family Artiviridae. Boxes indicate open reading frames as annotated on GenBank accession AB555544 with the addition of −1 frameshifting (Zhai et al., 2010). |
Biology
Artivirids have been described in several arthropods including insects and crustaceans. Virus isolates have been obtained from Culex (Isawa et al., 2011) and Armigeres subalbatus (Zhai et al., 2010) mosquitoes, as well as shrimp (Poulos et al., 2006). High-throughput sequencing has further described artiviruses in thrips (Thekke-Veetil et al., 2020), Drosophila (Wu et al., 2010), and in other diptera, fleas, ants, and crabs (Shi et al., 2016). Infections in insects are believed to be persistent, however artiviruses can cause a fatal disease in shrimp (Poulos et al., 2006).
Derivation of names
Artiviridae, Artivirus: from arthropod totivirus, referring to the hosts of members in the family and the relationship to totiviruses; the suffix viridae for family taxa or the suffix -virus for genus taxa. Species epithets are Latinized Japanese numbers (Figure 2 Artiviridae).
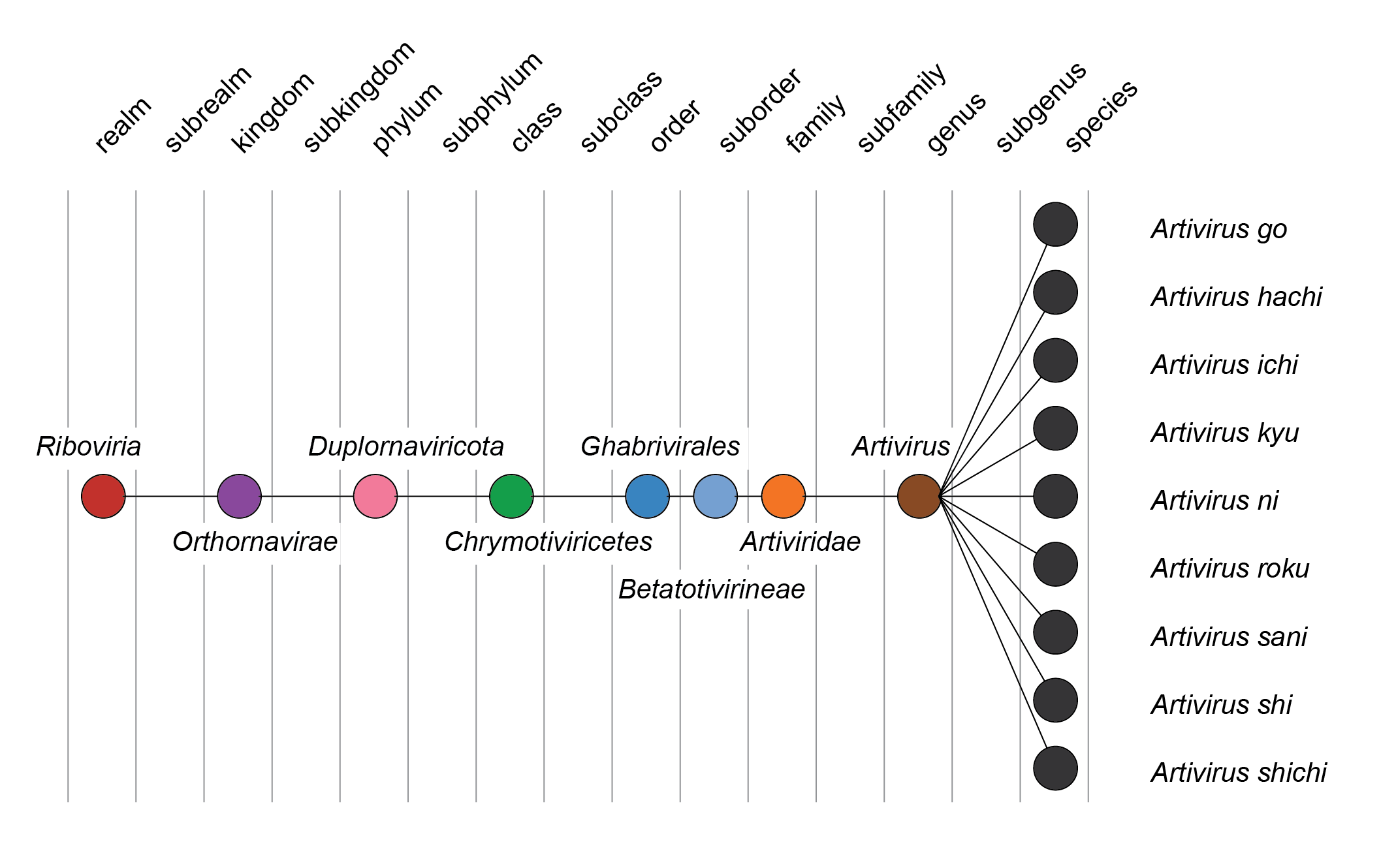 |
| Figure 2 Artiviridae. Taxonomy of the family Artiviridae. |
Species demarcation criteria
1) The host species for the virus must be assigned convincingly.
2) For viral genome sequences assembled from high-throughput sequencing reads, and not validated in full by Sanger sequencing, read numbers, RPKM values, and/or coverage depth values for the assembled sequence must be specified, or the read data must be available at NCBI such that these values can be determined.
3) Virus genome sequences that share <70% RdRP amino acid sequence identity will be considered to represent different virus species regardless of whether reported from the same or different host species.
4) Virus genome sequences that share ![]() 70% RdRP amino acid sequence identity, but are reported from different host species, are considered to represent the same virus species.
70% RdRP amino acid sequence identity, but are reported from different host species, are considered to represent the same virus species.

