Family: Polycipiviridae
Ingrida Olendraite, Katherine Brown, Steven M. Valles, Andrew E. Firth, Yanping Chen, Diego M. A. Guérin, Yoshifumi Hashimoto, Salvador Herrero, Joachim R. de Miranda and Eugene Ryabov
The citation for this ICTV Report chapter is the summary published as Olendraite et al., (2019):
ICTV Virus Taxonomy Profile: Polycipiviridae, Journal of General Virology, 100, 554–555.
Corresponding author: Andrew E. Firth ([email protected])
Edited by: Nick J. Knowles and Sead Sabanadzovic
Posted: February 2019
PDF: ICTV_Polycipiviridae.pdf
Summary
Polycipiviridae is a family of picorna-like viruses with non-segmented, linear, positive-sense RNA genomes of approximately 10–12 kb (Table 1. Polycipiviridae). Unusually for viruses within the order Picornavirales, their genomes are polycistronic, with four (or more) consecutive 5′-proximal open reading frames (ORFs) encoding structural (and possibly other) proteins and a long 3′ ORF encoding the replication polyprotein. Members of species within the family have all been derived from arthropods, with the majority coming from ants.
Table 1. Polycipiviridae. Characteristics of members of the family Polycipiviridae.
| Characteristic | Description |
| Typical member | Solenopsis invicta virus 2 (MF041813), species Sopolycivirus solenopsis |
| Virion | Thought to be non-enveloped, 33 nm in diameter |
| Genome | 10–12 kb of positive-sense, non-segmented RNA |
| Replication | Not studied; presumed to be similar to other Picornavirales members |
| Translation | Directly from genomic RNA, presumed internal ribosome entry site elements in 5′-untranslated region and intergenic region |
| Host range | Arthropoda |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Pisuviricota, class Pisoniviricetes, order Picornavirales; includes the genera Chipolycivirus, Hupolycivirus and Sopolycivirus, a total of 14 species |
Virion
Morphology
Icosahedral particles approximately 33 nm in diameter have been observed by electron microscopy in samples prepared from ants infected with Solenopsis invicta virus 2 (SINV2) but not in samples prepared from non-infected ants and these particles are assumed to represent SINV2 virions (Valles et al., 2007) (Figure 1. .Polycipiviridae). Genomes are picorna-like and encode three jelly-roll fold capsid proteins, thus particles are assumed to be picorna-like, i.e. non-enveloped, icosahedral, pseudo-T=3 symmetry comprising 60 copies of each of the ORF1, ORF3 and ORF4 products. However, this has not yet been experimentally confirmed.
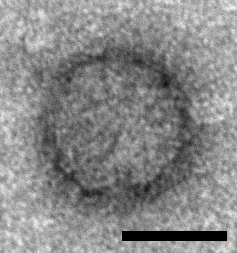 |
| Figure 1. Polycipiviridae. Electron micrograph of a virus-like particle purified from Solenopsis invicta worker ants infected with Solenopsis invicta virus 2 (scale bar = 20 nm) [reproduced from (Valles et al., 2007); U.S. government material as public domain content]. |
Physicochemical and physical properties
Virion physicochemical properties have not been investigated.
Nucleic acid
The genome is thought to comprise a single positive-sense RNA molecule of 10–12 kb. The genome has a 3′-poly(A) tail and is presumed to have a small viral protein genome-linked (VPg) covalently attached to the 5′-end.
Proteins
The virion protein composition has not been experimentally determined.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genome contains four main 5′-proximal ORFs (ORFs 1–4) and one long 3′ ORF (ORF5) (Figure 2. Polycipiviridae) (Olendraite et al., 2017). ORF1, ORF3 and ORF4 encode products with homology to picornavirus jelly-roll fold capsid proteins. ORF2 encodes a product of unknown function. ORF5 encodes superfamily III helicase, chymotrypsin-like serine protease and superfamily I RNA dependent RNA polymerase (RdRP) domains, and is presumed to encode a viral protein genome-linked (VPg) and potentially another protein between the helicase and protease, and one or two additional proteins upstream of the helicase. ORF5 is expected to be proteolytically cleaved by the viral protease but the cleavage sites have not been mapped. The ORFs are flanked by 5′- and 3′-untranslated regions (UTRs) and a lengthy intergenic region (IGR) between ORF4 and ORF5. The 5′-UTR and intergenic region (IGR) are presumed to contain internal ribosome entry site (IRES) elements to direct translation of ORF1 and ORF5 whereas ORFs 2–4 have been proposed to be expressed via a ribosome re-initiation mechanism. Members of the genus Sopolycivirus contain an additional ORF (2b) overlapping the 5′-end of ORF2; ORF2b encodes a small protein with a predicted transmembrane domain. Members of at least one species (Formica exsecta virus 3) contain a further additional small ORF inserted between ORF2 and ORF3. Replication has not been studied but is assumed to be similar to other members of the order Picornavirales.
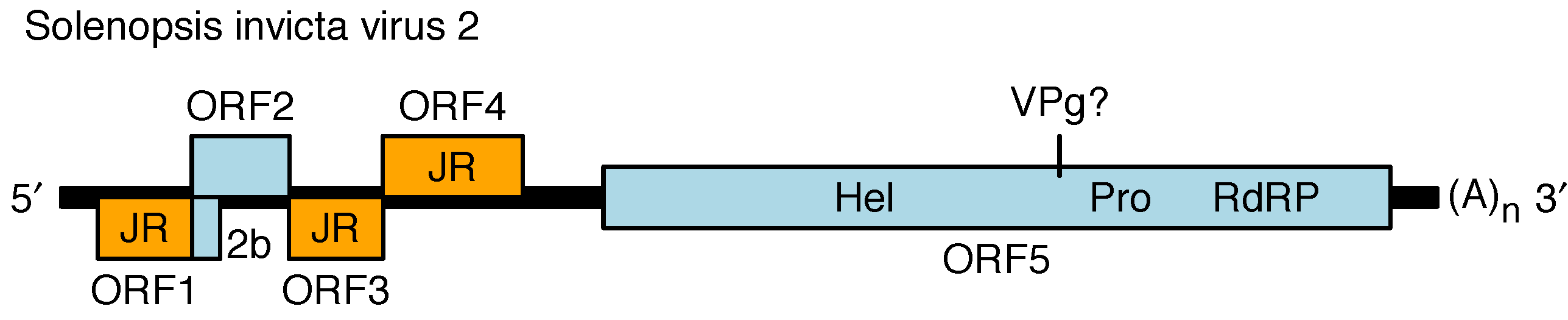 |
| Figure 2. Polycipiviridae Genome organization of Solenopsis invicta virus 2 (genus Sopolycivirus). The 5′-proximal ORFs encode jelly-roll capsid protein domains (JR, orange), a protein of unknown function (ORF2), and a small predicted transmembrane protein (2b; specific to genus Sopolycivirus). The 3′ ORF encodes helicase (Hel), protease (Pro), and RNA-dependent RNA polymerase (RdRP) domains. The genome is polyadenylated and believed to have a viral protein genome-linked (VPg) covalently attached to its 5′-end. Members of the genera Hupolycivirus and Chipolycivirus have similar genome organizations except that they lack the 2b ORF. |
Biology
All current family members derive from ants or from transcriptomic studies of arthropods. Members of the type species Solenopsis invicta virus 2 appear to establish a chronic, asymptomatic infection in Solenopsis invicta ants and high virus loads have been detected in larvae, pupae, workers and queens (Hashimoto and Valles 2008a, Hashimoto and Valles 2008b). In infected ants, 96% of SINV2 genome equivalents localize to the midgut section of the alimentary canal. Further, SINV2 has been experimentally transmitted to non-infected ants via feeding suggesting food-borne transmission probably by trophallaxis (Hashimoto and Valles 2008b). Fire ant infections with SINV2 are associated with negative impacts on queen ants resulting in significant reductions in fecundity, longer claustral periods, and slower growth of newly established colonies (Manfredini et al., 2016).
Derivation of names
Polycipi: derived from polycistronic picorna-like.
Genus demarcation criteria
Genera are separated based on phylogenetic clustering based on protein sequence identities (Figure 3. Polycipiviridae). However formal genus demarcation criteria have not yet been established.
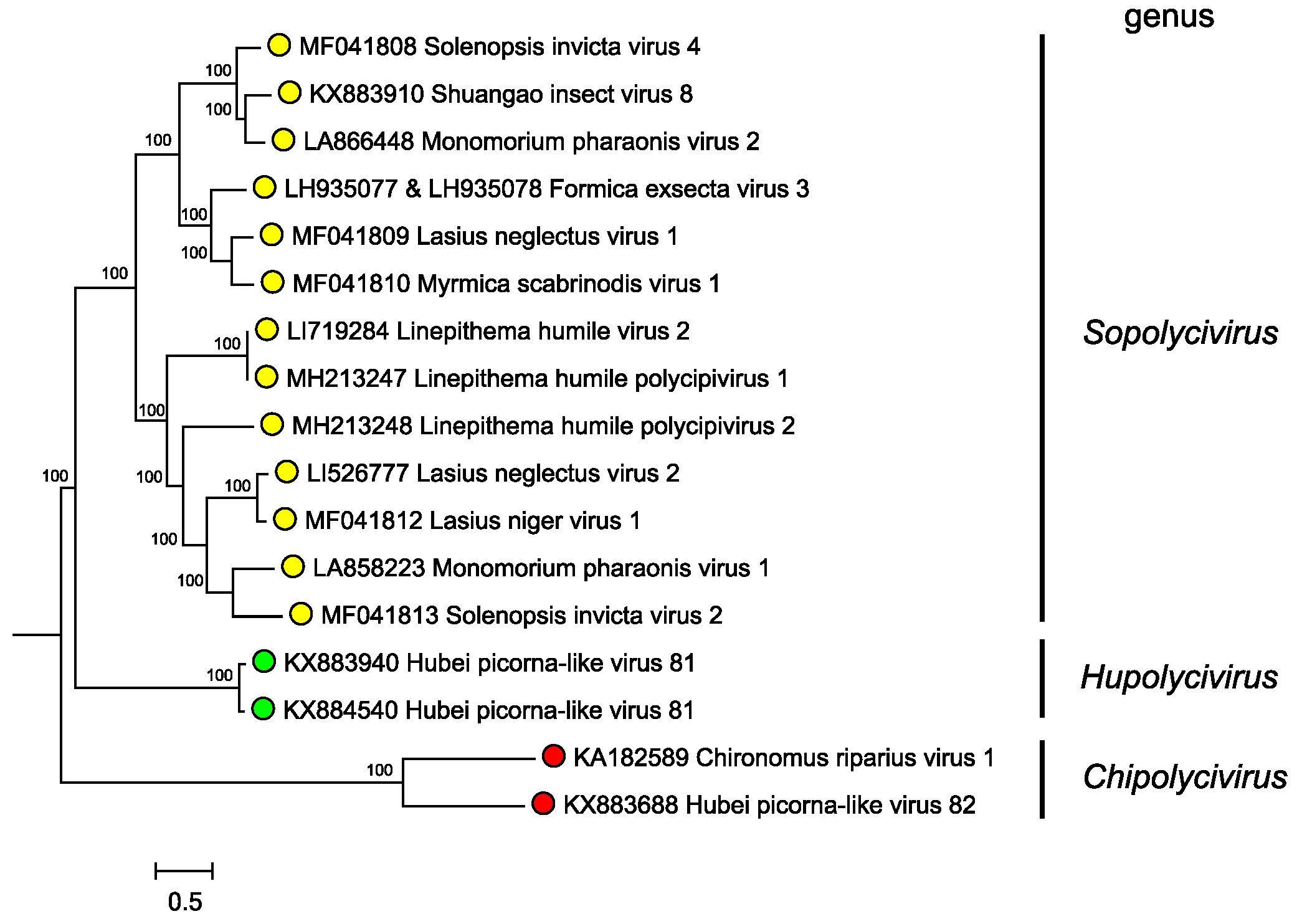 |
| Figure 3. Polycipiviridae. Mid-point rooted phylogenetic tree constructed from full-length ORF5 amino acid sequences of members of polycipivirus species and related unclassified virus sequences. Genera are annotated at left. Sequences were aligned with MUSCLE, and a Bayesian Markov chain Monte Carlo-based phylogenetic tree (ngen=1,000,000) was produced with MrBayes. Posterior probabilities are indicated at the branching points. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships within the family
Phylogenetic analysis of the ORF5 non-structural polyprotein amino acid sequences indicates that sopolyciviruses, hupolyciviruses and chipolyciviruses form distinct monophyletic groups in the family (Figure 3. Polycipiviridae) (Olendraite et al., 2017). Monophyly of the Polycipiviridae family within the order Picornavirales is not completely certain (Aiewsakun and Simmonds 2018), though the unusual genome organization of these viruses justifies their current taxonomic grouping.
Relationships with other taxa
Similar to other members of the order Picornavirales, polycipiviruses have polyadenylated positive-sense RNA genomes that encode three jelly-roll capsid proteins, a superfamily III helicase, a 3C-like chymotrypsin-related protease and a superfamily I RdRP (Olendraite et al., 2017). Unlike other members of the order, with the exception of marnaviruses, the polycipivirus protease has serine rather than cysteine at the active site. Polycipiviruses also have a unique genome organization (Figure 2. Polycipiviridae).

