Family: Pistolviridae (Interim Report)
This is a summary page created by the ICTV Report Editors using information from associated Taxonomic Proposals and the Master Species List.
Edited by: Holly Hughes
Posted: October 2024
Summary
The family Pistolviridae includes dsRNA viruses of vertebrates (Table 1 Pistolviridae). The family Pistolviridae was established in 2024 (Master Species List #39). The family is comprised of one genus, Pistolvirus, and four species.
Table 1 Pistolviridae. Characteristics of members of the family Pistolviridae.
| Characteristic | Description |
| Example | piscine myocarditis-like virus (KT725636), species Pistolvirus ni, genus Pistolvirus |
| Virion | Non-enveloped, icosahedral particles, 50 nm in diameter |
| Genome | A linear dsRNA of 5.8–6.7 kbp with three ORFs (Figure 1 Pistolviridae) |
| Replication | Unknown |
| Translation | RdRP possibly expressed by −1 ribosomal frameshifting |
| Host range | Vertebrates |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Duplornaviricota, class Chrymotiviricetes, order Ghabrivirales: 1 genus and 4 species (Figure 2 Pistolviridae) |
Virion
Morphology
Pistolviruses have been shown to be icosahedral and non-enveloped, about 50nm in diameter. The virion is suggested to have structural symmetry and surface projections (Haugland et al., 2011).
Nucleic acid
The nucleic acid is dsRNA of 5.8–6.7 kb with three open reading frames (ORF). The two upstream ORFs are in an overlapping frame, whereas the third downstream ORF is in a non-overlapping frame (Haugland et al., 2011).
Proteins
The upstream ORF1 produces the capsid protein (about 91 kDa), the overlapping ORF2 produces the RNA-dependent RNA polymerase (RdRP, about 83 kDa), and the downstream ORF3 produces a hypothetical protein (about 33kDa) that has a C-X-C chemokine motif (Mor and Phelps 2016).
Genome organization and replication
The replication strategy of pistolviruses is not yet fully elucidated. Virions have been shown to be internalized into cells, presumably after binding an unknown receptor (Haugland et al., 2011). Virus accumulation in cell culture supernatant following uptake into cells was observed suggesting virion exocytosis (Haugland et al., 2011). The translation of the capsid and RdRP is the product of a fusion protein following a −1 ribosomal frameshift resulting from the presence of a pseudoknot structure in the intergenic region (Sandlund et al., 2021) (Figure 1 Pistolviridae).
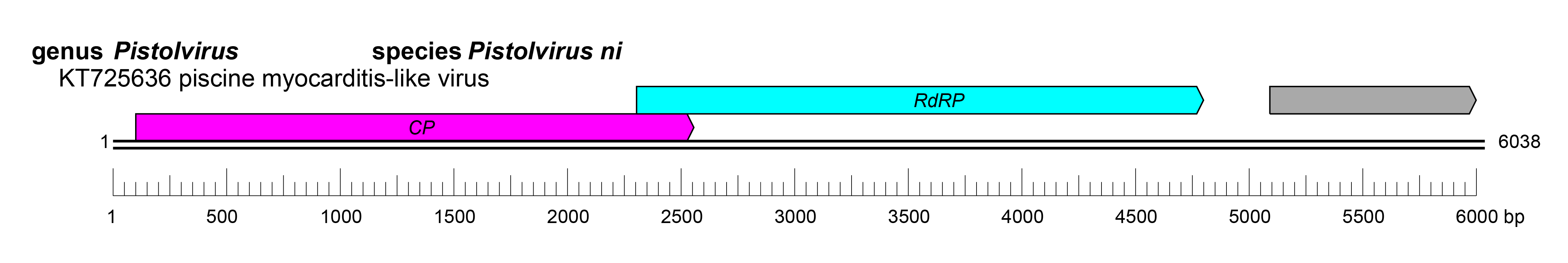 |
| Figure 1 Pistolviridae. Genome organisation of Piscine myocarditis-like virus, a member of the family Pistolviridae. Boxes indicate open reading frames as annotated on GenBank accession KT725636. |
Biology
Pistolvirids have been described in fish and are unique among ghabrivirals as these viruses are detected in vertebrate hosts. Piscine myocarditis virus has been isolated and implicated in myocarditis in Atlantic salmon (Salmo salar) where horizontal transmission of the virus was noted (Haugland et al., 2011). In contrast, pistolviruses can also be detected in healthy fish, where piscine myocarditis-like virus was isolated from golden shiner bait fish (Notemigonus crysoleucas) (Mor and Phelps 2016). Additional pistolviruses have been described via high-throughput sequencing of carp (Cyprinus carpio) and lump sucker (Cyclopterus lumpus) (Sandlund et al., 2021).
Derivation of names
Pistolviridae, Pistolvirus: from piscine toti-like, referring to the hosts of viruses in the family and the relationship to totiviruses; the suffix viridae for family taxa or the suffix -virus for genus taxa. Epithets are derived from Latinized Japanese numbers (Figure 2 Pistolviridae).
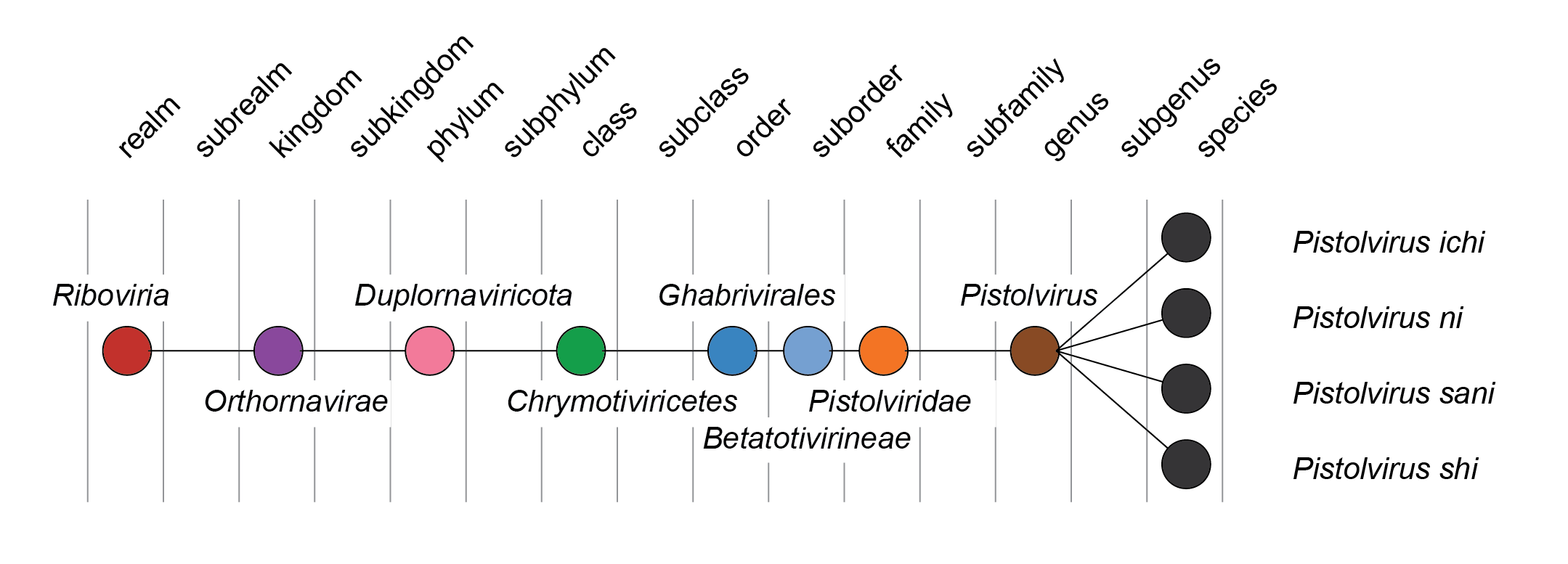 |
| Figure 2 Pistolviridae. Taxonomy of the family Pistolviridae. |
Species demarcation criteria
1) The host species for the virus must be assigned convincingly.
2) For viral genome sequences assembled from high-throughput sequencing reads, and not validated in full by Sanger sequencing, read numbers, RPKM values, and/or coverage depth values for the assembled sequence must be specified, or the read data must be available at NCBI such that these values can be determined.
3) Virus genome sequences that share <70% RdRP amino acid sequence identity will be considered to represent different virus species regardless of whether reported from the same or different host species.
4) Virus genome sequences that share ![]() 70% RdRP amino acid sequence identity, but are reported from different host species, are considered to represent the same virus species.
70% RdRP amino acid sequence identity, but are reported from different host species, are considered to represent the same virus species.

