Family: Yueviridae
Mart Krupovic, Yuri I. Wolf, Eugene V. Koonin, and Jens H. Kuhn
The citation for this ICTV Report chapter is the summary published as:
Corresponding authors: Jens H. Kuhn ([email protected])
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: September 2023
Summary
Yueviridae is a family of negative-sense RNA viruses with genomes of 7.8–8.2 kb (Table 1.Yueviridae). These viruses have been associated with crustaceans, insects, stramenopiles, and plants. The family includes a single genus with two species for two viruses. The yuevirid genome consists of two segments, each with at least one open reading frame (ORF). The large (L) segment ORF encodes a large protein containing an RNA-directed RNA polymerase domain. The small (S) segment ORF encodes a nucleocapsid protein.
Table 1.Yueviridae Characteristics of members of the family Yueviridae
| Characteristic | Description |
| Example | Běihǎi sesarmid crab virus 3 (L: KX884032; S: KX884033), species Yuyuevirus beihaiense, genus Yuyuevirus |
| Virion | Unknown |
| Genome | 7.8–8.2 kb of bi-segmented negative-sense RNA |
| Replication | Unknown |
| Translation | Unknown |
| Host range | Branchiopod, decapod, and isopod crustaceans, dipteran and thysanopteran insects, lycopodiopsid and polypodiopsid plants and various macrophytes, and stramenopiles |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Yunchangviricetes, order Goujianvirales; the family includes one genus and two species |
Virion
Morphology
Unknown
Nucleic acid
Yuevirids have two segments of linear negative-sense RNA with a total length of 7.8–8.2 kb (large (L) segment: 6.6–6.9 kb; small (S) segment: 1.2–1.3 kb) (Shi et al., 2016).
Genome organization and replication
Viruses of the family Yueviridae have bi-segmented genomes, with each segment possessing at least one ORF (Figure 1.Yueviridae). The L segment ORF (L) encodes a large protein containing an RNA-directed RNA polymerase (RdRP) with a GDP:polyribonucleotidyltransferase (PRNTase) domain that is related to that of viruses in the order Mononegavirales, suggesting a similar capping mechanism. The S segment ORF (N) encodes a nucleocapsid protein structurally related to the homologs encoded by mononegavirals, in particular paramyxovirids. The replication cycle of yuevirids remains to be elucidated.
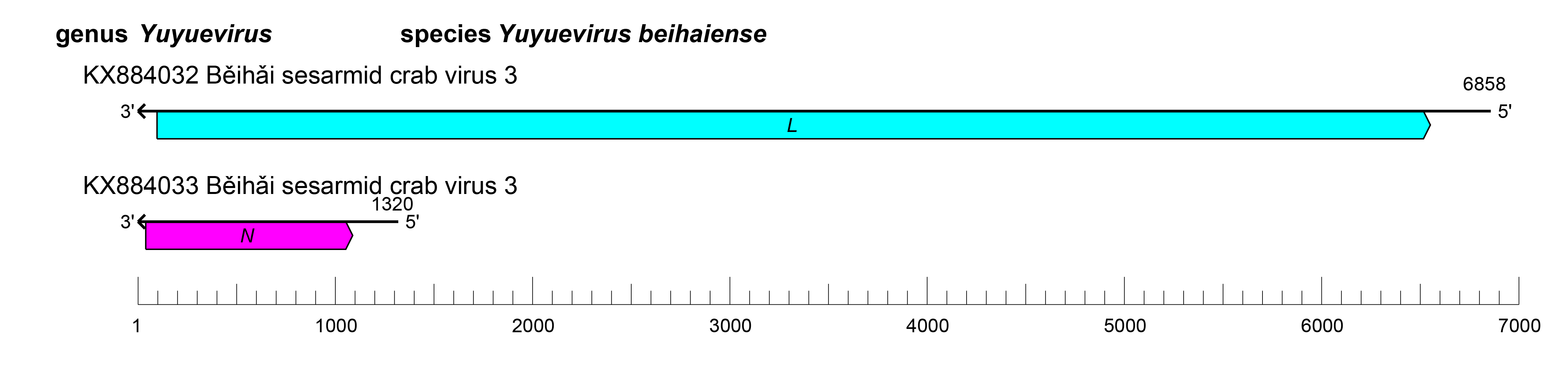 |
| Figure 1.Yueviridae. Genome length, organization, and position of open reading frames (ORFs) of a representative yuevirid, Běihǎi sesarmid crab virus 3. ORFs are indicated as boxes, colored according to the predicted protein function (L, large protein gene encoding an RNA-directed RNA polymerase [RdRP] domain; N, gene encoding a nucleocapsid protein). |
Biology
The only two classified yuevirids, Běihǎi sesarmid crab virus 3 (BhSCV3) and Shāhé yuèvirus-like virus 1 (ShYLV1), have been found in unspecified sesarmid crabs (Crustacea: Decapoda) and freshwater isopods (Crustacea: Isopoda), respectively, in China (Shi et al., 2016).
Unclassified yuevirids have been associated with brine shrimp of various species (Xuan et al., 2023), stramenopiles (bacillariaceaen Pseudo-nitzschia heimii Manguin, 1957 and downy mildews (peronosporaceaen Bremia lactucae Regel, 1843 and Plasmopara viticola (Berk. & M.A. Curtis) Berl. & De Toni, 1888)) (Chiapello et al., 2020, Charon et al., 2021, Chase et al., 2021), and onion thrips (thripid Thrips tabaci Lindeman, 1889) in Italy (Chiapello et al., 2021); mosquitoes (culicid Ochlerotatus hexodontus Dyar, 1916) in Finland (Truong Nguyen et al., 2022); and meadow spikemoss (selaginellaceaen Selaginella apoda ((L.) Spring)) (Mifsud et al., 2022), threeleaf goldback ferns (pteridaceaen Pityrogramma trifoliata L.) (Mifsud et al., 2022), and unspecified macrophytes in the USA (Rosario et al., 2022). The diversity of yuevirids is likely vastly underestimated (Käfer et al., 2019, Hou et al., 2023, Olendraite et al., 2023).
Derivation of names
beihaiense: from Běihǎi sesarmid crab virus 3
shaheense: from Shāhé yuèvirus-like virus 1
Yueviridae: from Yuè (越), an ancient Chinese state during the Spring and Autumn Period
Yuyuevirus: from the Chinese Yúyuè (於越), a synonym for Yuè (越), an ancient Chinese state during the Spring and Autumn Period
Genus demarcation criteria
Not applicable (the family includes only a single genus).
Species demarcation criteria
Members of different species are phylogenetically distinct based upon analysis of the RdRP. Numerical demarcation values have not been established.
Relationships within the family
Phylogenetic relationships of members of the family Yueviridae are shown in Figure 2.Yueviridae.
 |
| Figure 2.Yueviridae. Phylogenetic relationships of yuevirids. The list of complete exemplar genomes of Negarnaviricota was obtained from the ICTV Virus Metadata Resource (VMR) datasheet v.38.1 (https://ictv.global/vmr). RdRP core domains were identified by running a PSI-BLAST search against either the protein coding gene complement or six-frame translation of the virus RNA sequences using the 143 negarnaviricot profiles (Neri et al., 2022) and a tosovirid profile (Waltzek et al., 2022). RdRP core fragments were clustered using MMSEQS2 with the sequence similarity threshold of 0.3; sequences in each cluster were aligned using MUSCLE5. A consensus sequence was derived from each alignment; consensus sequences were aligned using MUSCLE5, then each consensus amino acid was expanded into an alignment column (Neri et al., 2022). An approximate ML phylogenetic tree was reconstructed from this alignment of 1,122 negarnaviricot RdRP core sequences using FastTree (WAG evolutionary model, gamma-distributed site rates). A representative sequence (one, closest to the family root) was selected for each of the 36 negarnaviricot families and the full tree was trimmed to these 36 representative leaves. |
Relationships with other taxa
Yuevirids are most closely related to other viruses in subphylum Haploviricotina, such as aspivirids, jingchuvirals, mononegavirals, and qinvirids.
Related, unclassified viruses
Additional viruses are described in (Xuan et al., 2023), but their accession numbers are not yet publicly available.
| Virus name | Accession number | Virus abbreviation | Reference |
| Bremia lactucae associated yuevirus-like virus 2 | L: MZ926716* | (Mifsud et al., 2022) | |
| Enontekio yuevirus | L: ON955264* | (Truong Nguyen et al., 2022) | |
| freshwater macrophyte associated yue-like virus 1 | L: ON125143* | (Rosario et al., 2022) | |
| Goldenrod fern yue-like virus | L: OW528586* | ||
| insect yue-like virus 1 | S: MW297844* | (Chiapello et al., 2021) | |
| meadow spikemoss associated yue-like virus | L: OW521095* | (Mifsud et al., 2022) | |
| Plasmopara viticola lesion associated Yue-like virus 1 | L: MN585286* | (Chiapello et al., 2020) | |
| Plasmopara viticola lesion associated Yue-like virus 2 | L: MN585287* | (Chiapello et al., 2020) | |
| Plasmopara viticola lesion associated Yue-like virus 3 | L: MN585288* | (Chiapello et al., 2020) | |
| Susy yue-like virus | L: BK059855* | (Charon et al., 2021) | |
| Thrips tabaci associated yue-like virus 1 | L: MN764157* | (Chiapello et al., 2021) |
Virus names and virus abbreviations are not official ICTV designations.
* Incomplete genome

