Family: Sunviridae
Jens H. Kuhn, Gael Kurath, Yuri I. Wolf, and Timothy H. Hyndman
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Timothy H. Hyndman ([email protected])
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: September 2023
Summary
Sunviridae is a family of negative-sense RNA viruses with genomes of about 17.2 kb (Table 1.Sunviridae). These viruses have been isolated from snakes in Australia. The family includes a single genus with one species for one virus. The genomes of sunvirids consist of nonsegmented RNA with six open reading frames (ORFs) that encode at least six proteins.
Table 1.Sunviridae. Characteristics of members of the family Sunviridae
| Characteristic | Description |
| Example | Sunshine Coast virus (JN192445), species Sunshinevirus reptilis genus Sunshinevirus |
| Virion | Enveloped, spherical |
| Genome | About 17.2 kb of nonsegmented negative-sense RNA |
| Replication | Unknown |
| Translation | Unknown |
| Host range | Squamate reptiles (pythonid and boid snakes) |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Monjiviricetes, order Mononegavirales. The family includes one genus and one species. |
Virion
Morphology
Infectivity is lost following treatment with chloroform. Consequently, it is assumed that sunvirids produce enveloped particles (Hyndman 2012). The envelopes are likely derived from host cell membranes and likely contain peplomers made of virus-encoded glycoproteins.
Nucleic acid
Sunvirids have nonsegmented negative-sense RNA with a total length of about 17.2 kb (Hyndman 2012, Hyndman et al., 2012a). Virions likely contain a single genome molecule. The RNA likely has a 3′-terminal free hydroxyl group and a 5′-triphosphate and is likely not polyadenylated. The ends of the RNA are speculated to have inverted complementary sequences encoding transcription and replication initiation signals. Complementary full-length RNA constitutes the replication intermediate; both genomic and antigenomic RNA is hypothesized to be tightly bound to nucleocapsid proteins (N).
Genome organization and replication
The genomes of sunvirids have nonsegmented genomes with six ORFs that are > 1kb which encode six proteins: nucleocapsid protein (N), phosphoprotein (P), matrix protein (M), fusion protein (F), glycoprotein (G), and a large (L) protein containing an RNA-directed RNA polymerase (RdRP) domain (Hyndman 2012, Hyndman et al., 2012a) (Figure 1.Sunviridae). The attachment protein is referred to as G, since phenotypic evidence of hemagglutination or neuraminidase activity was not detected with Sunshine Coast virus (Hyndman 2012). A 450-nucleotide ORF (nominally referred to as “X”) exists between the N and P ORFs but it is not known whether this is transcribed and translated. The replication cycle of sunvirids remains to be determined. In snake cell culture analysis, sunvirids caused cytopathic effects in the form of syncytia (Hyndman et al., 2012a).
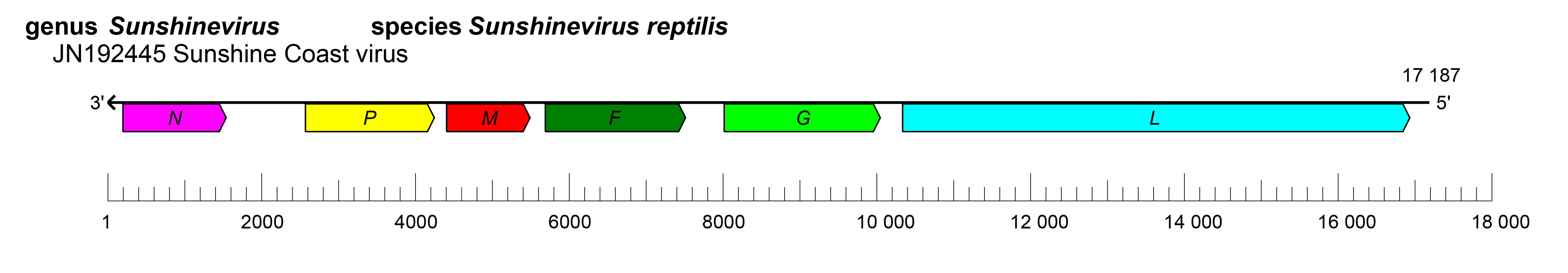 |
| Figure 1.Sunviridae. Genome length, organization, and position of open reading frames (ORFs) of Sunshine Coast virus, the only classified virus in family Sunviridae. ORFs are indicated as boxes, colored according to the predicted protein function (N, nucleocapsid protein gene; P, phosphoprotein gene; M, matrix protein gene; F, fusion protein gene; G, glycoprotein gene; L, large protein gene). |
Biology
Sunshine Coast virus (SunCV; previously also known as Sunshine virus), has been isolated from black-headed pythons (pythonid Aspidites melanocephalus (Krefft, 1864)), jungle carpet pythons (pythonid Morelia spilota (Lacépède, 1804)), spotted pythons (pythonid Antaresia maculosa (Peters, 1873)), and woma pythons (pythonid Aspidites ramsayi (Macleay, 1882)) in Australia (Hyndman 2012, Hyndman et al., 2012a, Hyndman et al., 2012b). Beyond lethargy, inappetance and occasional bronchointerstitial pneumonia, SunCV causes neurorespiratory disease in snakes. The virus has been detected in a number of organs but appears to be neurotropic; pathological changes associated with infection are most often detected in brain tissue and typically consist of white matter spongiosis of the hindbrain in the absence of gross lesions (Hyndman et al., 2012b). Infected carpet pythons tend to shed virus persistently, and latent infections are not uncommon. SunCV is found in the mouths and cloacas of infected snakes, suggesting virus transmission via oral and/or cloacal secretions. Virus RNA has been detected in embryos and allantois and amnion membranes of infected carpet python dams, but positive embryos were not viable, suggesting inefficient or self-limiting vertical transmission (Hyndman et al., 2012b, Hyndman and Johnson 2015).
A distinct, SunCV-related unclassified virus was associated with a respiratory disease outbreak among captive sidewinders (viperid Crotalus cerastes Crotalus cerastes) in a North American zoo (Wellehan et al., 2020); no nucleotide sequence is available for this virus.
Derivation of names
reptilis: from Latin reptilis, meaning “reptile”
Sunshinevirus: from Sunshine Coast virus and -virus: suffix for a virus genus
Sunviridae: from Sunshine Coast virus and -viridae: suffix for a virus family
Genus demarcation criteria
Not applicable (the family includes only a single genus).
Species demarcation criteria
Not applicable (the only genus includes only a single species).
Relationships within the family
A coding-complete genome sequence is available for only one virus isolate.
Relationships with other taxa
Sunvirids are most closely related to mononegaviral filovirids, paramyxovirids, and pneumovirids (Hyndman et al., 2012a) (Figure 2.Sunviridae).
 |
| Figure 2.Sunviridae. Phylogenetic relationships of Sunviridae. The list of complete exemplar genomes of Negarnaviricota was obtained from the ICTV Virus Metadata Resource (VMR) datasheet v.38.1 (https://ictv.global/vmr). RdRP core domains were identified by running a PSI-BLAST search against either the protein coding gene complement or six-frame translation of the virus RNA sequences using the 143 negarnaviricot profiles (Neri et al., 2022) and a tosovirid profile (Waltzek et al., 2022). RdRP core fragments were clustered using MMSEQS2 with the sequence similarity threshold of 0.3; sequences in each cluster were aligned using MUSCLE5. A consensus sequence was derived from each alignment; consensus sequences were aligned using MUSCLE5, then each consensus amino acid was expanded into an alignment column (Neri et al., 2022). An approximate ML phylogenetic tree was reconstructed from this alignment of 1,122 negarnaviricot RdRP core sequences using FastTree (WAG evolutionary model, gamma-distributed site rates). A representative sequence (one, closest to the family root) was selected for each of the 36 negarnaviricot families and the full tree was trimmed to these 36 representative leaves. |
Related, unclassified viruses
| Virus | Accession number | Abbreviation | Reference |
| sidewinder-associated sunshine-like virus | (Wellehan et al., 2020); |

