Family: Narnaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Viruses in the family Narnaviridae consist of a single molecule of non-encapsidated positive-strand RNA of 2.3–2.9 kb, which encodes a single protein of 80–104 kDa with amino acid sequence motifs characteristic of an RNA dependent RNA polymerase (RdRp).
Genus Narnavirus
Type species Saccharomyces 20S RNA narnavirus
Virion properties
Morphology
No true virions are found associated with members of this genus. The genomes, however, are associated with their RdRps forming ribonucleoprotein complexes in 1:1 stoichiometry. Genetic and biochemical evidence show that they are located in the cytoplasm.
Physicochemical and physical properties
The ribonucleoprotein complex sediments through a sucrose gradient with a sedimentation coefficient of about 20S. These complexes are quite stable at pH 9.0 and have in vitro RNA polymerase activity that synthesizes mainly 20S RNA, and a minor amount of complementary strands.
Nucleic acid
The Saccharomyces 20S RNA narnavirus (ScNV-20S) genome is a linear ssRNA of 2.5 kb in size with a high G+C content (ca. 60%). There is no poly(A) tail at the 3’ end and it is not known whether the 5’ end is capped. It is present in a high copy number under stress conditions, such as growth under nitrogen starvation, reaching up to 100,000 copies/cell.
Proteins
No structural proteins have been described for members of this family. ScNV-20S has a coding capacity for a protein of 91 kDa (p91), with sequences conserved among RdRps. The conserved sequences are more similar to those of replicases of ssRNA enterobacteria phages than polymerases of members of the family Totiviridae in the same host. This protein is quite basic (estimated pI of 11) and has ssRNA binding activity. Protein p91 is essential for replication and responsible for the in vitro RdRp activity that synthesizes 20S RNA. P91 does not undergo proteolytic processing after translation. Studies using antibodies against this protein show that it is expressed in yeast cells grown exponentially or under induction conditions.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
ScNV-20S has one ORF that encodes p91, and there are no ORFs with coding capacity larger than 100 aa in the complementary strand. The ORF for p91 spans almost the entire sequence of 20S RNA, with a short untranslated leader sequence at the 5’ end (12 nt) and an UTR at the 3’ end of 12 nt (Figure 1). Two replication models for 20S RNA have been proposed based on the similarity of p91 to the replicases of RNA enterobacteria phages and the replication intermediates obtained in the in vitro RNA polymerase reaction. One model is similar to the replication cycle of ssRNA enterobacteria phages such as Qβ; that is, ScNV-20S is copied into the complementary strands and these copies serve as templates for 20S RNA synthesis. Annealing of 20S RNA and its complementary strand gives a double-stranded form of ScNV-20S. This dsRNA called W can be easily isolated from all ScNV-20S-containing yeast strains. The other model hypothesizes that W dsRNA is the replicative form of ScNV-20S. At present, available data support the first model. Recently, a reverse genetics system for ScNV-20S has been established. Like native viruses, viruses generated from cDNA vectors can be transmitted to daughter cells indefinitely without the vector or any selection.
Antigenic properties
No antibody has been raised from virus particle preparations.
Biological properties
ScNV-20S infects more than 90% of laboratory strains of the baker’s yeast Saccharomyces cerevisiae. Some strains isolated from the brewery industry also have been found to carry ScNV-20S. There is no phenotype associated with the presence of this RNA. Like other viruses of fungi, there is no extracellular stage in the ScNV-20S life cycle. Transmission takes place through mating or cytoplasmic mixing. These viruses are very stable. Known curing procedures that eliminate members of the family Totiviridae in the same host, such us growth at high temperature, or addition of either cycloheximide, acridine orange, or guanidine HCl, do not eliminate ScNV-20S.
Species demarcation criteria in the genus
Narnaviruses generally replicate stably within the cell as the cells grow. Virus strains of the same species are expected to segregate relative to each other as the cells grow, whereas those of different species should be stably co-maintained. Viruses of the same species should be similarly affected by host chromosomal mutations. Viruses that can recombine or exchange segments with each other to give viable progeny should be considered the same species. Although these biological criteria are the prime determinants of species, sequence criteria also are used. Less than 50% sequence identity at the protein level generally reflects a species difference. None of the above criteria is absolute, but narnaviruses described so far leave little doubt about species demarcation. For example, ScNV-20S and ScNV-23S are only 30% identical in the 439 aa region of highest similarity. More important, they are stably compatible with each other in the same yeast strain.
List of species in the genus Narnavirus
| Saccharomyces 20S RNA narnavirus |
|
|
| Saccharomyces 20S RNA narnavirus 37-4C | [AF039063] | (ScNV-20S-37-4C) |
| Saccharomyces 23S RNA narnavirus |
|
|
| Saccharomyces 23S RNA narnavirus 37-4C | [U90136] | (ScNV-23S-37-4C) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Narnavirus but have not been approved as species
None reported.
Genus Mitovirus
Type species Cryphonectria mitovirus 1
Virion properties
Morphology
No true virions are found associated with members of this genus. Genetic and biochemical evidence shows that they are located in the mitochondria.
Physicochemical and physical properties
Mitochondrial fractions isolated in sucrose gradients contain mitovirus ds- and ssRNA. No subfractionation of mitochondria has been achieved.
Nucleic acid
The virus genome consists of a single molecule of RNA of 2.3–2.7 kb. Double-stranded RNAs in this size range can be isolated from mitochondria of infected isolates. Single-stranded RNA of the same size, and corresponding to the coding strand of the dsRNA, is present in infected tissue in greater molar amount than the dsRNA. The 5′ and 3′ sequences can be folded into stable stem-loop structures. For some mitoviruses, the 5′ and 3′ sequences are complementary. The coding strand has 62–73% A+U residues, but no poly(A) tail is associated with the 3′ end.
Proteins
No structural proteins are known to be associated with the virus ssRNA or dsRNA.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The putative coding strand is predicted to be translatable only in mitochondria, not in the cytoplasm. When mitochondrial codon usage is invoked (UGA coding for tryptophan), the deduced translation product is a protein of 80–97 kDa, containing RdRp motifs. RdRp activity and an 80 kDa RdRp protein have been detected in mitochondria from an infected Ophiostoma novo-ulmi isolate. No large polypeptide is predicted from the complementary strand of any mitovirus.
Antigenic properties
No antibody has ever been raised from virus particle preparations.
Biological properties
Mitoviruses have been found in isolates of the chestnut blight fungus, Cryphonectria parasitica, Dutch elm disease fungi, Ophiostoma novo-ulmi and O. ulmi, and Sclerotinia homoeocarpa, the cause of dollar spot of turf grass. Fungal isolates may contain one or several mitoviruses. Some, but not all, member viruses reduce virulence of the fungus (i.e., cause "hypovirulence"). Mitoviruses are localized in mitochondria. They can be transmitted to uninfected strains by hyphal fusion (anastomosis). The transmission rate through asexual spores (conidia) is virus-specific and varies from 10 to 100%. In C. parasitica, transmission through sexual spores (ascospores) occurs at 20–50% when the infected parent is the female in matings, but does not occur when the infected parent is male in matings. In O. novo-ulmi, viruses are usually excluded from ascospores, even when both parents are infected. Identical mitoviruses have been found in O. novo-ulmi and O. ulmi, and a strain of Ophiostoma mitovirus 3a has been reported in Sclerotinia homoeocarpa, suggesting that both interspecies and intergenus virus transmission occurs in nature.
Species demarcation criteria in the genus
Species demarcation criteria have not been precisely defined. However, amino acid sequence identities of putative RdRp proteins between the different mitovirus species so far defined are less than 40%. Amino acid sequence identities of putative RdRp proteins between strains of the same mitovirus species are greater than 90%.
List of species in the genus Mitovirus
| Cryphonectria mitovirus 1 |
|
|
| Cryphonectria mitovirus 1 cpNB631 | [L31849] | (CMV-1-cpNB631) |
| Ophiostoma mitovirus 3a |
|
|
| Ophiostoma mitovirus 3a OnuLd | [AJ004930] | (OMV-3a-OnuLd) |
| Sclerotinia homoeocarpa mitovirus | [AY172454] |
|
| Ophiostoma mitovirus 4 |
|
|
| Ophiostoma mitovirus 4 OnuLd | [AJ132754] | (OMV-4-OnuLd) |
| Ophiostoma mitovirus 5 |
|
|
| Ophiostoma mitovirus 5 OnuLd | [AJ132755] | (OMV-5-OnuLd) |
| Ophiostoma mitovirus 6 |
|
|
| Ophiostoma mitovirus 6 OnuLd | [AJ132756] | (OMV-6-OnuLd) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Mitovirus but have not been approved as species
| Botrytis mitovirus 1 | [EF580100] | (BMV-1) |
| Gremmeniella mitovirus S1 | [AF534641] | (GMV-S1) |
| Helicobasidium mitovirus 1 | [AB110977] | (HMV-1) |
| Ophiostoma mitovirus 1a | [AM087548] | (OMV-1a) |
| Ophiostoma mitovirus 1b | [AM087549] | (OMV-1b) |
| Ophiostoma mitovirus 2 |
| (OMV-2) |
| Ophiostoma mitovirus 3b | [AM087550] | (OMV-3b) |
| Thielaviopsis mitovirus 1 | [AY563138] | (TMV-1) |
List of other related viruses which may be members of the family Narnaviridae but have not been approved as species
| Rhizoctonia virus M2 | [U51331] | (RVM2) |
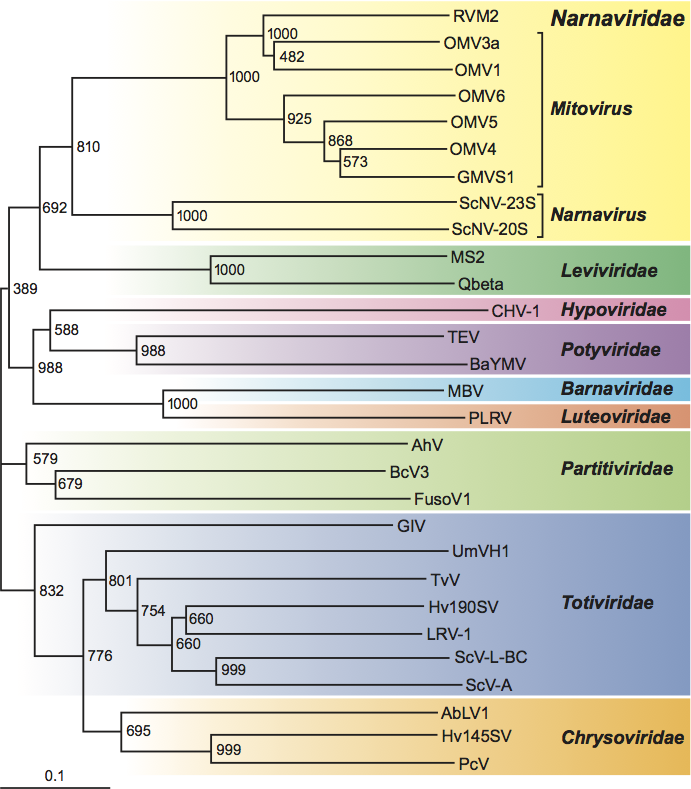
Phylogenetic relationships within the family
In a neighbor-joining phylogenetic tree based on aa sequences of the putative RdRp proteins, the mitovirus and narnavirus genera are clearly distinguished, but nevertheless form a significant cluster (Figure 2). The putative RdRp protein of the unassigned virus, Rhizoctonia virus M2 (RVM2), clusters with those of the mitoviruses (Figure 2). However, since only a small proportion of RVM2 co-purifies with mitochondria with most of it being found in the cytoplasm, RVM2 does not use the mitochondrial code. Furthermore, there is evidence for a RVM2 DNA copy in the host genome. These findings suggest significant differences between RVM2 and the mitoviruses.
Similarity with other taxa
The putative RdRp proteins of narnaviruses and mitoviruses are distantly related to those of bacteriophages in the family Leviviridae (Figure 2). Furthermore, the 3’-end secondary structures of members of the genus Narnavirus resemble those of coliphages in the family Leviviridae. In a neighbor-joining phylogenetic tree of families of fungus viruses and related viruses in other taxa, based on aa sequences of the putative RdRp proteins, the families Narnaviridae and Leviviridae form a cluster with 69.2% bootstrap support (Figure 2).
Derivation of names
Narna: from naked RNA virus.
Mito: from mitochondrial.
Further reading
Buck and Brasier, 2002 K.W. Buck, C.M. Brasier, S.M. Tavantzis, Viruses of the Dutch elm disease fungiDsRNA Genetic Elements: Concepts and Applications in Agriculture, Forestry and Medicine. In: S.M. Tavantzis, DsRNA Genetic Elements: Concepts and Applications in Agriculture, Forestry and Medicine. CRC Press, Boca Raton, FL2002165–190.
Esteban and Fujimura, 2003 R. Esteban, T. Fujimura, Launching the yeast 23S RNA narnavirus shows 5’ and 3’ cis-acting signals for replication. Proc. Natl Acad. Sci., U S A. 100 (2003) 2568–2573.
Hong et al., 1998 Y. Hong, T.E. Cole, C.M. Brasier, K.W. Buck, Evolutionary relationships among putative RNA-dependent RNA polymerases encoded by a mitochondrial virus-like RNA in the Dutch elm disease fungus, Ophiostoma novo-ulmi, by other viruses and virus-like RNAs and by the Arabidopsis mitochondrial genome. Virology. 246 (1998) 158–169.
Polashock and Hillman, 1994 J.J. Polashock, B.I. Hillman, A small mitochondrial double-stranded (ds) RNA element associated with a hypovirulent strain of the chestnut blight fungus and ancestrally related to yeast cytoplasmic T and W dsRNAs. Proc. Natl Acad. Sci., U S A. 91 (1994) 8680–8684.
Polashock et al., 1997 J.J. Polashock, P.J. Bedker, B.I. Hillman, A mitochondrial dsRNA of Cryphonectria parasitica: Ascospore inheritance and mitochondrial recombination. Mol. Gen. Genet. 256 (1997) 566–571.
Rodríguez-Cousiño et al., 1991 N. Rodríguez-Cousiño, L.M. Esteban, R. Esteban, Molecular cloning and characterization of W double-stranded RNA, a linear molecule present in Saccharomyces cerevisiae: identification of its single-stranded RNA form as 20S RNA. J. Biol. Chem. 266 (1991) 12772–12778.
Solórzano et al., 2000 A. Solórzano, N. Rodríguez-Cousiño, R. Esteban, T. Fujimura, Persistent yeast single-stranded RNA viruses exist in vivo as genomic RNA:RNA polymerase complexes in 1:1 stoichiometry. J. Biol. Chem. 275 (2000) 26428–26435.
Tavantzis et al., 2002 S.M. Tavantzis, . Lakshman, C. Liu, S.M. Tavantzis, Double-stranded RNA elements modulating virulence in Rhizoctonia solaniDsRNA Genetic Elements: Concepts and Applications in Agriculture, Forestry and Medicine. In: S.M. Tavantzis, DsRNA Genetic Elements: Concepts and Applications in Agriculture, Forestry and Medicine. CRC Press, Boca Raton, FL2002191–211.
Widner et al., 1991 W.R. Widner, Y. Matsumoto, R.B. Wickner, Is 20S RNA naked?. Mol. Cell. Biol. 11 (1991) 2905–2908.
Wu et al., 2010 M. Wu, L. Zhang, G. Li, D. Jiang, S.A. Ghabrial, Genome characterization of a debilitation-associated mitovirus infecting the phytopathogenic fungus Botrytis cinerea. Virology. 406 (2010) 117–126.
Contributed by
Hillman, B.I. and Esteban, R.
Figures
Figure 1 Genomic organization of Saccharomyces 20S RNA narnavirus (ScNV-20S) and Saccharomyces 23S RNA narnavirus (ScNV-23S) and the proteins encoded on them (p91 and p104, respectively). Sequence motifs (A to D) conserved in RdRp are boxed and shaded. Motifs 1, 2 and 3 are present only in p91 and p104.
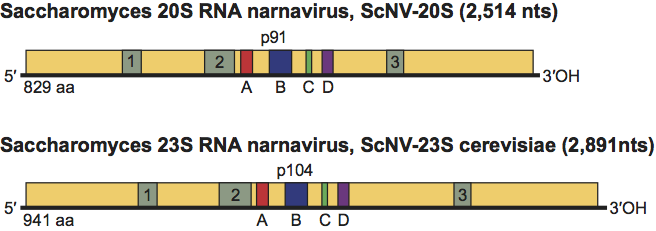
Figure 2 Phylogenetic tree based on aa sequences of motifs A to E (Hong et al. 1998) of the putative RdRp proteins of members of the family Narnaviridae, other families of RNA viruses of fungi and related viruses in other host taxa, and the family Leviviridae of RNA bacteriophages. Sequence alignments and the neighbor-joining tree were made using the Clustal X program. Bootstrap numbers (1000 replicates) are shown on the nodes. Abbreviations and sequence acquisition numbers are: AbVL1, Agaricus bisporus virus L1 X94361; AhV, Atkinsonella hypoxylon virus L39126; BaYMV, Barley yellow mosaic virus D01091; BcV3, Beet cryptic virus 3 S63913; CHV1, Cryphonectria hypovirus 1 M57938; CMV1, Cryphonectria mitovirus 1 L31849; FusoV1, Fusarium solani virus 1 D55668; GlV, Giardia lamblia virus L13218; GMVS1, Gremmeniella mitovirus S1 AF534641; Hv145SV, Helminthosporium victoriae 145S virus AF297176; Hv190SV, Helminthosporium victoriae 190S virus U41345; LRV1, Leishmania RNA virus 1-1 M92355; MBV, Mushroom bacilliform virus U07551; MS2, Enterobacteria phage MS2 GB-PH:MS2CG; OMV3a, Ophiostoma mitovirus 3a AJ004930; OMV4, Ophiostoma mitovirus 4 AJ132754; OMV5, Ophiostoma mitovirus 5 AJ132755; OMV6, Ophiostoma mitovirus 6 AJ132756; PcV, Penicillium chrysogenum virus AF296439; PLRV, Potato leafroll virus X14600; Qbeta, Enterobacteria phage Q AY099114; RVM2, Rhizoctonia virus M2 U51331; ScV-L-A, Saccharomyces cerevisiae virus L-A J04692; ScV-L-BC, Saccharomyces cerevisiae virus L-BC U01060; ScNV-20S, Saccharomyces 20S RNA narnavirus M63893; ScNV-23S, Saccharomyces 23S RNA narnavirus M86595; TEV, Tobacco etch virus M15239; TvV, Trichomonas vaginalis virus U08999; UmVH1, Ustilago maydis virus H1 U01059.