Genus: Varicosavirus
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Distinguishing features
Plant viruses with two components of negative sense ssRNA and non-enveloped flexuous rod-shaped virions. They are only distantly related to other ssRNA viruses such as the Rhabdoviridae.
Virion properties
Morphology
Virions are fragile non-enveloped rods mostly measuring 320–360×about 18 nm; each has a central canal about 3 nm in diameter and an obvious helix with a pitch of about 5 nm (Figure 1). As virions are very unstable in vitro, their detection and visualization may be facilitated by prior fixation with glutaraldehyde. The helix of particles, especially those in purified preparations, tends to loosen and particles are then seen as partially uncoiled filaments.
Physicochemical and physical properties
Virus particles have a density in Cs2SO4 of about 1.27 g cm−3.
Nucleic acid
The genome of lettuce big-vein associated virus (LBVaV) consists of ssRNA, but after deproteinization of purified virions, both ds- and ssRNA are detected. The genome is 12.9 kb in size, divided into two components of 6.8 kb (RNA-1) and 6.1 kb (RNA-2); their 3′-termini have no poly(A) tracts. The 3′- and 5′-terminal sequences of the two RNAs are similar, but do not exhibit inverse complementarities. Conserved motifs in the transcription start and end signals resemble those of viruses in the order Mononegavirales.
Proteins
The size of the coat protein, when estimated by PAGE, is about 48 kDa. The CP cistron, located on RNA-2, encodes a protein of 397 aa with a predicted size of 44.5 kDa.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The first genomic segment (RNA-1) contains one small putative ORF possibly coding for a 5 kDa protein and one large ORF that encodes a protein of 232 kDa (designated L protein by analogy with the L polymerase of rhabdoviruses) (Figure 2). The second genomic segment (RNA-2) contains five ORFs. The first ORF encodes the coat protein (44.5 kDa) and the second to the fifth ORFs code for proteins with unknown functions (36, 32, 19 and 41 kDa respectively) (Figure 2). Although the genome organization of LBVaV is similar to that of rhabdoviruses, LBVaV does not have a non-coding leader sequence. The aa sequence of the L protein is homologous with the L polymerases of some negative sense RNA viruses, especially those of rhabdoviruses.
Antigenic properties
The CP is a rather poor antigen. LBVaV and tobacco stunt virus (TStV) are serologically closely related.
Biological properties
Host range
Varicosaviruses occur naturally in two families of plant species (Compositae and Solanaceae). LBVaV and TStV infect some common experimental host species.
Transmission
Both LBVaV and TStV are transmitted in soil and in hydroponic systems by zoospores of the chytrid fungus Olpidium brassicae. The non-crucifer strain of O. brassicae transmitting Mirafiori lettuce big-vein virus (MLBVV; genus Ophiovirus) and TStV in Japan has recently been referred to as Olpidium virulentus and the new name was also used for other O. brassicae isolates infecting lettuce and shown to be associated with the presence of LBVaV and MLBVV in Australia. A further proposal based on genetic and host range differences of Olpidium spp. from many countries suggested that lettuce-infecting isolates could be named Olpidium compositae.
Due to the longevity of infectious LBVaV from stored O. brassicae resting spores, it is assumed that the virus is carried internally by the fungus.
The viruses are also transmitted experimentally, sometimes with difficulty, by mechanical inoculation. Neither of the viruses is reported to be seed-transmitted.
Cytopathic effects
None reported.
Species demarcation criteria in the genus
Not yet definable.
List of species in the genus Varicosavirus
| Lettuce big-vein associated virus |
|
|
| Lettuce big-vein associated virus-Japan:Kagawa | [AB075039=NC_0011558+AB114138=NC_011568] | (LBVaV-JP) |
| Tobacco stunt virus | [AB190521*] | (TStV) |
Species names are in italic script; isolate names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequence does not comprise the complete genome.
List of other related viruses which may be members of the genus Varicosavirus but have not been approved as species
TStV was considered to be distinct from LBVaV. However, based on nucleotide and amino acid sequence identities between the two, the former was later considered to be a tobacco-infecting strain of the latter.
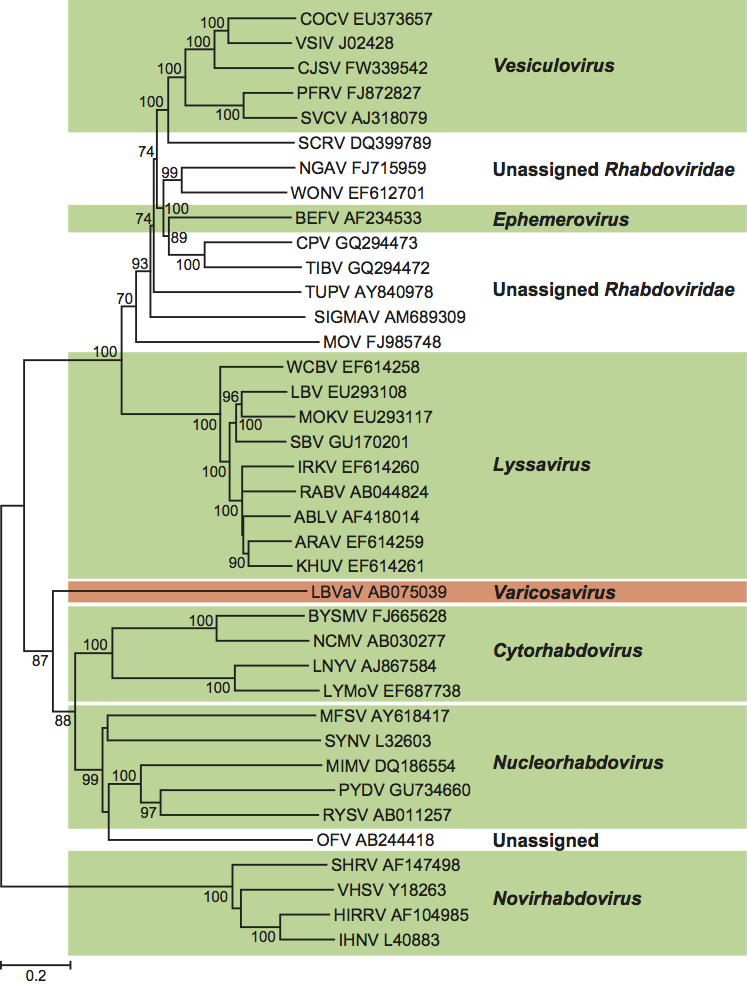
Phylogenetic relationships within the genus
Comparison of the CP coding regions of eight LBVaV isolates from Japan and Spain and five TStV isolates from Japan showed they were identical in size and had nucleotide and amino acid sequence identities of 95.6–96.5% and 97.2–98.7%, respectively. This and a comparison of sequences of the other genes of one TStV isolate with those of LBVaV led to the conclusion that TStV was a strain of LBVaV. A later, larger analysis of the CP coding regions of 29 LBVaV isolates (from Europe, Australia and Japan) and the five TStV isolates from Japan, revealed sequence identities of 93.6–99.7% (nucleotides) and 94.3–100% (amino acids); the nucleotide and amino acid sequence identities of the 29 LBVaV isolates alone had the same range. The identities of the five TStV were 95.7–99.7% (nucleotides) and 97.2–100% (amino acids). This was taken to support TStV being a strain of LBVaV. In both analyses, all five TStV isolates grouped together in a clade of their own and there were four amino acid differences unique to the TStV isolates. This and the fact LBVaV isolates do not infect tobacco and TStV isolates do not infect lettuce might suggest some merit in keeping an open mind on the strain/species issue until further evidence is available. (See Figure 3.)
Similarity with other taxa
The genome structure and probable transcription mechanism of LBVaV indicates that it has a close relationship with rhabdoviruses (Figure 3). The aa sequences of both the CP and the L protein of LBVaV have significant similarities with those of rhabdoviruses. LBVaV also resembles rhabdoviruses in possessing conserved transcription termination/polyadenylation signal-like poly(U) tracts and in transcribing monocistronic RNAs. The presence of poly(U) tracts in the NCRs of LBVaV RNA-1 and RNA-2 suggest that transcription of LBVaV is regulated by a mechanism similar to that of rhabdoviruses (order Mononegavirales; family Rhabdoviridae). However, whereas rhabdoviruses contain a single negative sense ssRNA, LBVaV has two such RNAs.
Derivation of name
Vari: from Latin varix, meaning abnormal dilation or enlargement of a vein or artery and referring to the symptom previously thought to be induced by the type species. However, lettuce big-vein disease, although long thought to be induced by a virus previously designated lettuce big-vein virus, is now considered to be caused by isolates of the species Mirafiori lettuce big-vein virus (genus Ophiovirus). Viruses of this species are soil-borne and often occur in lettuce together with isolates of LBVaV.
Further reading
Hartwright et al., 2010 L.M. Hartwright, P.J. Hunter, J.A. Walsh, A comparison of Olpidium isolates from a range of host plants using internal transcribed spacer sequence analysis and host range studies. Fungal Biology. 114 (2010) 26–33.
Kuwata and Kubo, 1986 Kuwata, S. and Kubo, S. (1986). Tobacco stunt virus. AAB Descriptions of Plant Viruses No. 313, 4 pp.
Kuwata et al., 1983 S. Kuwata, S. Kubo, S. Yamashita, Y. Doi, Rod-shaped particles, a probable entity of lettuce big vein virus. Ann. Phytopath. Soc. Japan. 49 (1983) 246–251.
Maccarone et al., 2010 L.D. Maccarone, M.J. Barbetti, K. Sivasithamparam, R.A.C. Jones, Comparisons of the coat protein genes of Lettuce big-vein associated virus isolates from Australia with those of isolates from other continents. Arch. Virol. 155 (2010) 765–770.
Maccarone et al., 2010 L.D. Maccarone, M.J. Barbetti, K. Sivasithamparam, R.A.C. Jones, Molecular genetic characterization of Olpidium virulentus isolates associated with big-vein diseased lettuce plants. Plant Disease. 94 (2010) 563–569.
Nagase et al., 2004 M. Nagase, D. Kiyomitsu, T. Sasaya, H. Koganezawa, S. Kuwata, The complete nucleotide sequence of Tobacco stunt virus. Jpn J. Phytopathol. 70 (2004) 261.
Roggero et al., 2000 P. Roggero, M. Ciuffo, A.M. Vaira, G.P. Accotto, V. Masenga, R.G. Milne, An Ophiovirus isolated from lettuce with big-vein symptoms. Arch. Virol. 145 (2000) 2629–2642.
Sasaya et al., 2002 T. Sasaya, K. Ishikawa, H. Koganezawa, The nucleotide sequence of RNA-1 of Lettuce big-vein virus, genus Varicosavirus, reveals its relation to nonsegmented negative strand RNA viruses. Virology. 297 (2002) 289–297.
Sasaya et al., 2005 T. Sasaya, K. Ishikawa, S. Kuwata, H. Koganezawa, Molecular analysis of coat protein coding region of tobacco stunt virus shows that it is a strain of Lettuce big-vein virus in the genus. Varicosavirus. Arch. Virol. 150 (2005) 1013–1021.
Sasaya and Koganezawa, 2006 T. Sasaya, H. Koganezawa, Molecular analysis and virus transmission tests place Olpidium virulentus, a vector of Mirafiori lettuce big-vein virus and tobacco stunt virus, as a distinct species rather than a strain of Olpidium brassicae. J. Gen. Plant Pathol. 72 (2006) 20–25.
Contributed by
Walsh, J.A. and Verbeek, M.
Figures
Figure 1 Negative contrast electron micrograph of virions of an isolate of lettuce big-vein associated virus. The bar represents 100 nm.
(Courtesy J.A. Walsh and C.M. Clay.)
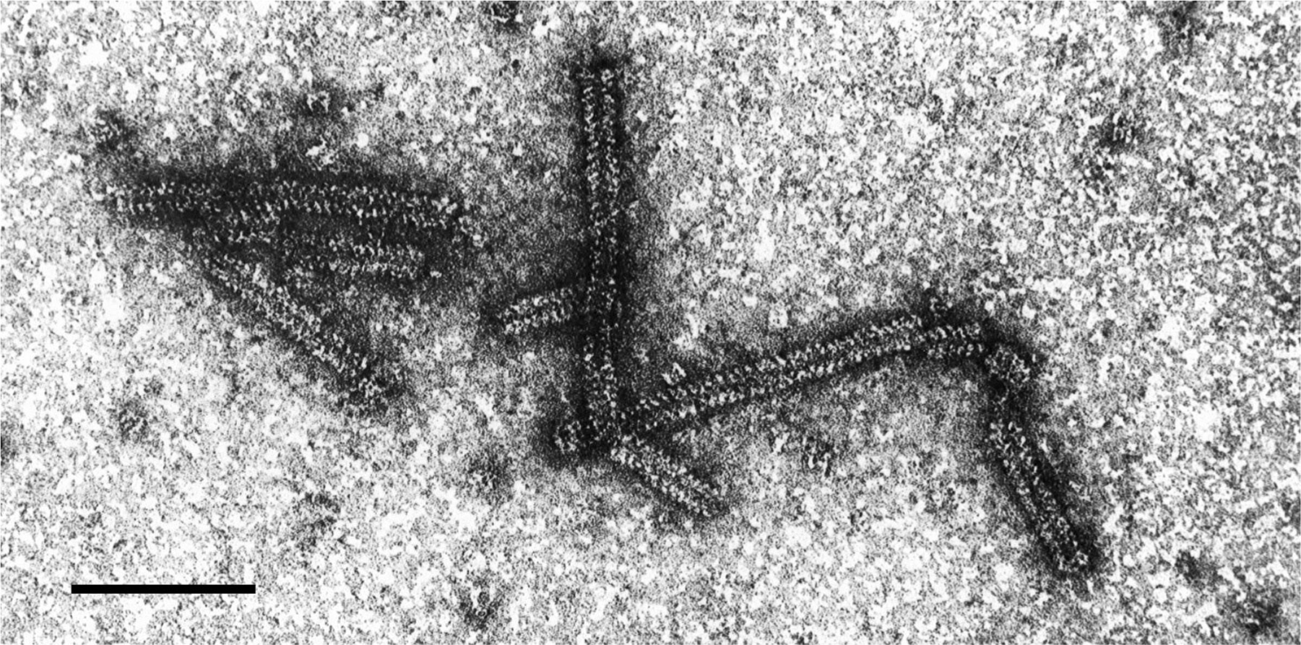
Figure 2 Diagram of the genome organization of the two genomic RNAs of lettuce big-vein associated virus. Solid lines represent RNA; boxes represent ORFs. ORFs are translated from the complementary strand vc.
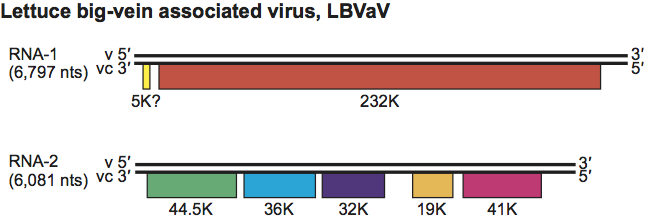
Figure 3 Phylogenetic tree based on the codon-aligned nucleotide sequences of the polymerase (L) genes of isolates of Lettuce big-vein associated virus, orchid fleck virus and members of the family Rhabdoviridae. The tree was prepared in MEGA4 using maximum composite likelihood distances and 10,000 bootstrap replicates (values shown where >60%). ABLV, Australian bat lyssavirus; ARAV, Aravan virus; BEFV, bovine ephemeral fever virus; BYSMV, barley yellow striate mosaic virus; CJSV, Carajas virus; COCV, Cocal virus; CPV, coastal plains virus; HIRRV, Hirame rhabdovirus; IHNV, infectious hematopoietic necrosis virus; IRKV, Irkut virus; KHUV, Khujand virus; LBV, Lagos bat virus; LBVaV, lettuce big-vein associated virus; LNYV, lettuce necrotic yellows virus; LYMoV, lettuce yellow mottle virus; MFSV, maize fine streak virus; MIMV, maize Iranian mosaic virus; MOKV, Mokola virus; MOV, Moussa virus; NCMV, northern cereal mosaic virus; NGAV, Ngaingan virus; OFV, orchid fleck virus; PFRV, pike fry rhabdovirus; PYDV, potato yellow dwarf virus; RABV, rabies virus; RYSV, rice yellow stunt virus; SBV, Shimoni bat virus; SCRV, Siniperca chuatsi rhabdovirus; SHRV, snakehead virus; SIGMAV, sigma virus; SVCV, spring viraemia of carp virus; SYNV, sonchus yellow net virus; TIBV, Tibrogargan virus; TUPV, Tupaia virus; VHSV, viral hemorrhagic septicemia virus; VSIV, vesicular stomatitis Indiana virus; WCBV, West Caucasian bat virus; WONV, Wongabel virus.
(Courtesy of Dr M.J. Adams; used by permission.)