Family: Bunyaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Morphological properties vary among viruses in each of the five genera; however, virions generally are spherical or pleomorphic, 80–120 nm in diameter, and display surface glycoprotein projections of 5–10 nm which are embedded in a lipid bilayered envelope approximately 5 nm thick. Virion envelopes are usually derived from cellular Golgi membranes, or on occasion, from cell surface membranes. Viral ribonucleocapsids are 2–2.5 nm in diameter, 200–3000 nm in length, and usually (but not always) display helical symmetry. (See Figure 1.)
Physicochemical and physical properties
The virion Mr is 300×106 to 400×106 and has an S20,W of 350–500. Virion buoyant densities in sucrose and CsCl are 1.16–1.18 and 1.20–1.21 g cm−3, respectively. Virions are sensitive to heat, lipid solvents, detergents and formaldehyde.
Nucleic acid
The viral genome comprises three unique molecules of negative or ambisense ssRNA, designated L (large), M (medium) and S (small), which total 11–19 kb (Table 1). The terminal nucleotides of each genome RNA segment are base-paired forming non-covalently closed, circular RNAs (and ribonucleocapsids). The terminal sequences of genome segments are conserved among viruses in each genus but are different from those of viruses in other genera. The genomic RNAs are not modified at their 5′ ends. The Mr of the genome ranges from 4.8×106 to 8×106 and this constitutes 1–2% of the virion by weight. Viral mRNAs are not polyadenylated and are truncated relative to the genomic RNAs at the 3′ termini. mRNAs have 5′-methylated caps and 10–18 non-templated nt at the 5′ end which are derived from host-cell mRNAs.
Table 1 Nucleotide lengths of selected completely sequenced genomes
| Genus | Virus | RNA segment | ||
| L | M | S | ||
| Orthobunyavirus |
|
|
| |
|
| Bunyamwera virus | 6875 | 4458 | 961 |
|
| California encephalitis virus - La Crosse virus | 6980 | 4526 | 980 |
| Hantavirus |
|
|
| |
| Hantaan virus - 76-118 | 6533 | 3616 | 1696 | |
| Seoul virus - HR80-39 | 6530 | 3651 | 1796 | |
| Puumala virus - Sotkamo | 6550 | 3682 | 1830 | |
| Sin Nombre virus - NMH10 | 6562 | 3696 | 2059 | |
| Nairovirus |
|
|
| |
| Dugbe virus | 12255 | 4888 | 1712 | |
| Crimean-Congo hemorrhagic fever virus (IbAr10200) | 12160 | 5366 | 1672 | |
| Phlebovirus |
|
|
| |
| Rift Valley fever virus | 6404 | 3884 | 1690 | |
| Sandfly fever Naples virus - Toscana virus | 6404 | 4215 | 1869 | |
|
| Uukuniemi virus | 6423 | 3231 | 1720 |
| Tospovirus |
|
|
| |
| Tomato spotted wilt virus | 8897 | 4821 | 2916 | |
| Impatiens necrotic spot virus | 8776 | 4972 | 2992 | |
Proteins
All viruses have four structural proteins, two external glycoproteins (Gn, Gc, named in accordance with their relative proximity to the amino or carboxy terminus of the polyprotein encoded by the M segment), a nucleocapsid protein (N) and a large (L) protein, an RNA-dependent RNA polymerase. Non-structural proteins are expressed from the S segments of some bunyaviruses, phleboviruses, tospoviruses and some hantaviruses, and from the M segments of bunyaviruses, nairoviruses, tospoviruses and some phleboviruses. Proteins encoded by each of the genome segments of viruses in each genus of the family are listed in Table 2.
Table 2 Deduced protein sizes (kDa)
| RNA | Protein | Genus | ||||
| Orthobunyavirus | Hantavirus | Nairovirus | Phlebovirus | Tospovirus | ||
| L segment |
|
|
|
|
| |
|
| L | 259–263 | 246–247 | 459 | 238–241 | 330–332 |
| M segment |
|
|
|
|
| |
| Gn | 29–41 | 68–76 | 30–45 | 50–70 | 46–58 | |
| Gc | 108–120 | 52–58 | 72–84 | 55–75 | 72–78 | |
| NSm | 15–18 | none | 78–85, 92–115 | none or 78 | 34 | |
| S segment |
|
|
|
|
| |
| N | 19–26 | 48–54 | 48–54 | 24–30 | 29 | |
| NSs | 10–13 | None or 7–12 | none | 29–31 | 52 | |
Lipids
Virions contain 20–30% lipids by weight. Lipids are derived from the membranes where viruses mature and include phopholipids, sterols, fatty acids and glycolipids.
Carbohydrates
Virions contain 2–7% carbohydrate by weight. Asparagine-linked sugars on the Gn and Gc proteins are largely of the high mannose type when viruses are grown in vertebrate cells.
Genome organization and replication
The genome organization of the different genera is shown in Figure 2. For all viruses, the L, M and S genome segments encode, respectively, the viral RNA polymerase (L protein), envelope glycoproteins (Gn and Gc) and nucleocapsid protein (N) in the virus-complementary sense RNA. The L protein is encoded in the complementary mRNA. A single, continuous ORF in the M RNA encodes the glycoproteins, and the primary gene product is co-translationally cleaved (except for nairoviruses) to give mature Gn and Gc. Hantaviruses and Uukuniemi-like phleboviruses encode no additional proteins in their M genome segments. Orthobunyaviruses and other phleboviruses encode a nonstructural protein (NSm) in the virion-complementary sense RNA. Nairoviruses encode two proteins of unknown functions: a mucin-rich protein and glycoprotein GP38, which are the products of posttranslational cleavage of preGn. Tospoviruses encode a NSm protein in an ambisense ORF at the 5′ end of virion-sense RNA. Some orthobunyaviruses and some hantaviruses encode a nonstructural protein (NSs) in an overlapping ORF to that encoding N in the 3′-half of the virion-sense S RNA. There is no direct evidence that nairoviruses encode any additional proteins in their S genome segments. Phleboviruses and tospoviruses encode a NSs protein in an ambisense ORF in the 5′-half of virion-sense S RNA. The NSs proteins of orthobunyaviruses, phleboviruses, and hantaviruses have been shown to act as interferon antagonists, NSs of tospoviruses as an RNAi antagonist. The NSm of phleboviruses can act as an apoptosis antagonist.
All stages of replication occur in the cytoplasm. The principal stages of replication are:
- Attachment, mediated by an interaction of one or both of the integral viral envelope proteins with, as yet unidentified, host receptors.
- Entry and uncoating, by endocytosis of virions and fusion of viral membranes with endosomal membranes.
- Primary transcription: i.e., the synthesis of mRNA species complementary to the genome templates by the virion-associated polymerase using host cell-derived capped primers (Figure 3).
- Translation of primary L and S segment mRNAs by free ribosomes; translation of M segment mRNAs by membrane-bound ribosomes and primary glycosylation of nascent envelope proteins. Co-translational cleavage of a precursor to yield Gn and Gc, and for some viruses, NSm.
- Synthesis and encapsidation of antigenome RNA to serve as templates for genomic RNA or, in some cases, for sgRNA.
- Genome replication (Figure 3).
- Secondary transcription; i.e., the amplified synthesis of the mRNA species and ambisense transcription.
- Morphogenesis, including accumulation of Gn and Gc in the Golgi, terminal glycosylation, acquisition of modified host membranes, generally by budding into the Golgi cisternae; budding at the cell surface has been observed with isolates of Rift Valley fever virus (genus Phlebovirus) in rat hepatocytes and Sin Nombre virus (genus Hantavirus) in polarized epithelial cells. RNPs of tomato spotted wilt virus (genus Tospovirus) are enwrapped by entire Golgi stacks leading to doubly enveloped virus particles. These fuse together and also with ER and lead to the formation of large ER-derived vesicles containing large amounts of mature, singly enveloped tospovirus particles.
- Fusion of cytoplasmic vesicles with the plasma membrane (except for tospoviruses) and release of mature virions.
Antigenic properties
One or both of the envelope glycoproteins display hemagglutinating and neutralizing antigenic determinants. Complement-fixing antigenic determinants are principally associated with nucleocapsid protein.
Biological properties
Viruses in the genera Orthobunyavirus, Nairovirus and Phlebovirus are capable of alternately replicating in vertebrates and arthropods, and generally are cytolytic for their vertebrate hosts, but cause little or no cytopathogenicity in their invertebrate hosts. Different viruses are transmitted by mosquitoes, ticks, phlebotomine flies, and other arthropod vectors. Some viruses display a very narrow host range, especially for arthropod vectors. No arthropod vector has been demonstrated for hantaviruses. Tospoviruses are transmitted by thrips and are capable of replicating in both thrips and plants. Transovarial and venereal transmission have been demonstrated for some viruses in their arthropod vectors. Aerosol infection occurs in certain situations or is the principal means of transmission for some viruses, particularly hantaviruses. In some instances, avian host and/or vector movements may result in virus dissemination. Some viruses cause a reduction in host-cell protein synthesis in vertebrate cells. Hantaviruses cause no detectable reduction in host macromolecular synthesis and routinely establish persistent, non-cytolytic infections in susceptible mammalian host cells, a finding consistent with their non-pathogenic persistence in their natural rodent or insectivore hosts. In natural infections of mammals, viruses are often targeted to a particular organ or cell type. Some viruses induce cell fusion at low pH. Some members have ion-dependent hemagglutinating activity. Genetic reassortment has been demonstrated for certain members both in vitro and in vivo.
Genus Orthobunyavirus
Type species Bunyamwera virus
Distinguishing features
The consensus terminal nucleotide sequences of the L, M and S genome segments are UCAUCACAUG… at the 3′ end and AGUAGUGUGC… at the 5′ end. The N and NSs proteins are encoded in overlapping reading frames by the S RNA and are translated from the same complementary mRNA as the result of alternate AUG initiation codon usage. Both glycoproteins and an NSm protein of 15–18 kDa are encoded as a precursor polyprotein by the M RNA. Genetic reassortment has been demonstrated between viruses, belonging to the same species but not between viruses from different species.
Viruses are serologically unrelated to members of other genera. Most viruses are mosquito-transmitted though some (e.g. the Tete group) are tick-transmitted. Occasionally alternate arthropods such as culicoid flies and phlebotomines transmit orthobunyaviruses. Some viruses are transmitted transovarially and venereally in arthopods.
Electron micrograph of negatively stained particles of California encephalitis virus strain La Crosse virus. The bar represents 100 nm.
(Courtesy of D. H. L. Bishop.)
Species demarcation criteria in the genus
The demarcation of orthobunyavirus species has proven difficult due to the lack of biochemical characterization of most of the named virus isolates. Species are thus primarily defined by serological criteria (cross-neutralization and cross-hemagglutination-inhibition tests). The limited available data indicate that one bunyavirus species is unable to form a reassortant with another species. Where known the aa sequences of the N proteins differ by more than 10%.
List of species in the genus Orthobunyavirus
| Acara virus |
|
|
|
| Acara virus - BeAn27639 | mosquitoes |
| (ACAV) |
| Moriche virus - TRVL57896 | mosquitoes |
| (MORV) |
| Akabane virus |
|
|
|
| Akabane virus - JaGAr39 | mosquitoes, culicoid flies | [S: M22011] | (AKAV) |
| Sabo virus - AN9398 | culicoid flies |
| (SABOV) |
| Tinaroo virus - CSIRO153 | culicoid flies | [S: AB000819] | (TINV) |
| Yaba-7 virus | N.D. |
| (Y7V) |
| Alajuela virus |
|
|
|
| Alajuela virus - 75V 2374 | mosquitoes |
| (ALJV) |
| Alajuela virus - 78V 2441 | mosquitoes |
| (ALJV) |
| San Juan virus - 75V 446 | mosquitoes |
| (SJV) |
| Anopheles A virus |
|
|
|
| Anopheles A virus – 1940 prototype | mosquitoes | [S: FJ660415] | (ANAV) |
| Anopheles A virus - CoAr3624 | mosquitoes |
| (ANAV) |
| Anopheles A virus - ColAn57389 | mosquitoes |
| (ANAV) |
| Las Maloyas virus - AG80-24 | mosquitoes |
| (LMV) |
| Lukuni virus - TRVL10076 | mosquitoes |
| (LUKV) |
| Trombetas virus- BeAn306770 | mosquitoes |
| (TRMV) |
| Anopheles B virus |
|
|
|
| Anopheles B virus - 1940 prototype | mosquitoes | [S: FJ660417] | (ANBV) |
| Boraceia virus - SPAr395 | mosquitoes | [S: FJ660418] | (BORV) |
| Bakau virus |
|
|
|
| Ketapang virus - MM2549 | mosquitoes |
| (KETV) |
| Bakau virus - MM2325 | mosquitoes |
| (BAKV) |
| Nola virus - DakArB2882 | mosquitoes |
| (NOLAV) |
| Tanjong Rabok virus - P9-87 | N.D. |
| (TRV) |
| Telok Forest virus - P72-4 | N.D. |
| (TFV) |
| Batama virus |
|
|
|
| Batama virus - AnB1292a | N.D. | [S: FJ660420] | (BMAV) |
| Benevides virus |
|
|
|
| Benevides virus - BeAn153564 | mosquitoes |
| (BVSV) |
| Bertioga virus |
|
|
|
| Bertioga virus - SPAn1098 | N.D. |
| (BERV) |
| Cananeia virus - SPAn64962 | mosquitoes |
| (CNAV) |
| Guaratuba virus – SAPAn12252 | mosquitoes |
| (GTBV) |
| Itimirim virus - SPAn47817 | N.D. |
| (ITIV) |
| Mirim virus - BeAn7722 | mosquitoes |
| (MIRV) |
| Bimiti virus |
|
|
|
| Bimiti virus - TRVL8362 | mosquitoes |
| (BIMV) |
| Botambi virus |
|
|
|
| Botambi virus - DakArB937 | mosquitoes |
| (BOTV) |
| Bunyamwera virus |
|
|
|
| Batai virus - MM2222 | mosquitoes | [S: X73464] | (BATV) |
| Birao virus - DakArB2198 | mosquitoes |
| (BIRV) |
| Bozo virus - ArB7343 | mosquitoes |
| (BOZOV) |
| Bunyamwera virus - 1943 prototype
| Mosquitoes
| [L: X14383; M: M11852; S: X73465] | (BUNV)
|
| Bunyamwera virus Laguna Larga - CbaAr426 | mosquitoes |
| (BUNV) |
| Cache Valley virus - 6V633 | mosquitoes | [S: X73465] | (CVV) |
|
|
|
|
|
| Fort Sherman virus - 86MSP18 | mosquitoes |
| (FSV) |
| Germiston virus - Ar1050 | mosquitoes | [M: M21951; S: M19420] | (GERV) |
| Iaco virus - BeAnr314206 | mosquitoes |
| (IACOV) |
| Ilesha virus - KO/2 | mosquitoes |
| (ILEV) |
| Lokern virus - FMS4332 | mosquitoes, culicoid flies |
| (LOKV) |
| Maguari virus - BeAr7272 | mosquitoes | [S: D00354] | (MAGV) |
| Maguari virus - AG83-1746 | mosquitoes |
| (MAGV) |
| Mboke virus - DakArY357 | mosquitoes |
| (MBOV) |
| Ngari virus - DakAr28542 | mosquitoes |
| (NRIV) |
| Northway virus - 0234 | mosquitoes | [S: X73470] | (NORV) |
| Playas virus - 75V3066 | mosquitoes |
| (PLAV) |
| Potosi virus | mosquitoes |
| (POTV) |
| Santa Rosa virus - M2-1493 | mosquitoes |
| (SARV) |
| Shokwe virus - SAAr4042 | mosquitoes |
| (SHOV) |
| Tensaw virus - A9-171b | mosquitoes |
| (TENV) |
| Tlacotalpan virus - 61D240 | mosquitoes |
| (TLAV) |
| Tucunduba virus- BeAr278 | mosquitoes |
| (TUCV) |
| Xingu virus- BeH388464 | mosquitoes? |
| (XINV) |
| Bushbush virus |
|
|
|
| Benfica virus - BeAn84381 | mosquitoes |
| (BNFV??) |
| Bushbush virus - TRVL26668 | mosquitoes |
| (BSBV) |
| Bushbush virus - GU71U 344 | mosquitoes |
| (BSBV) |
| Juan Diaz virus - MARU8563 | N.D. |
| (JDV) |
| Bwamba virus |
|
|
|
| Bwamba virus - M459 | mosquitoes |
| (BWAV) |
| Pongola virus - SAAr1 | mosquitoes |
| (PGAV) |
| California encephalitis virus |
|
|
|
| California encephalitis virus - BFS283 | mosquitoes | [S: U12797] | (CEV) |
| Inkoo virus - KN3641 | mosquitoes | [S: Z68496; M: U88059] | (INKV) |
| Jamestown Canyon virus - 61V-2235 | mosquitoes | [S: U12796; M: U88058] | (JCV) |
| Keystone virus - C14031-33 | mosquitoes | [S: U12801] | (KEYV) |
| La Crosse virus | mosquitoes | [L: U12396; M: D00202; S: K00610] | (LACV) |
| Lumbo virus | mosquitoes | [S: X73468] | (LUMV) |
| Melao virus-TRVL9375 | mosquitoes | [S: U12802; M: 88057] | (MELV) |
| Melao virus - AG83 497 | mosquitoes |
| (MELV) |
| San Angelo virus 20230 | mosquitoes | [S: U47139] | (SAV) |
| Serra do Navio virus - BeAr103645 | mosquitoes | [S: U47140] | (SDNV) |
| Snowshoe hare virus- Original | mosquitoes | [M: K02539; S: J02390] | (SSHV) |
| South River virus (Jamestown Canyon virus?) | mosquitoes | [S: U47141] | (SORV) |
| Tahyna virus- 92 | mosquitoes | [S: Z68497] | (TAHV) |
| Trivittatus virus- Original | mosquitoes | [S: U12803] | (TVTV) |
| Capim virus |
|
|
|
| Capim virus - BeAn 8582 | mosquitoes |
| (CAPV) |
| Caraparu virus |
|
|
|
| Apeu virus - BeAn848 | mosquitoes | [S: DQ188952; M: DQ188959] | (APEUV) |
| Bruconha virus- 77V-14814 | mosquitoes | [S: DQ188953] | (BRUV) |
| Caraparu virus - BeAn3994 | mosquitoes | [S: DQ188948; M: DQ188960; L: EF122411] | (CARV) |
| Ossa virus - BT1820 | mosquitoes | [S: DQ188954] | (OSSAV) |
| Vinces virus - 75V-807 | mosquitoes | [S: DQ188958; M: AF499012] | (VINV) |
| Catu virus |
|
|
|
| Catu virus - BeH151 | mosquitoes |
| (CATUV) |
| Estero Real virus |
|
|
|
| Estero Real virus - K329 | ticks |
| (ERV) |
| Gamboa virus |
|
|
|
| Gamboa virus - 75V 2621 | mosquitoes |
| (GAMV) |
| Gamboa virus - MARU10962 | mosquitoes |
| (GAMV) |
| Pueblo Viejo virus - E4-816 | mosquitoes |
| (PVV) |
| Guajara virus |
|
|
|
| Guajara virus – BeAn10615 | mosquitoes |
| (GJAV) |
| Guajara virus - GU71U 350 | mosquitoes |
| (GJAV) |
| Guama virus |
|
|
|
| Guama virus - BeAn 277 virus | mosquitoes |
| (GMAV) |
| Ananindeua virus - BeAn109303 | mosquitoes |
| (ANUV) |
| Moju virus - BeAr12590 | mosquitoes |
| (MOJUV) |
| Mahogany Hammock virus - FE4-2s | N.D. |
| (MGHV??) |
| Guaroa virus |
|
|
|
| Guaroa virus - 352111 | mosquitoes | [S: X73466] | (GROV) |
| Kairi virus |
|
|
|
| Kairi virus- TRVL8900 | mosquitoes | [S: X73467] | (KRIV) |
| Kaeng Khoi virus |
|
|
|
| Kaeng Khoi virus - S19-8 | nest bugs |
| (KKV) |
| Koongol virus |
|
|
|
| Koongol virus - MRM31 | mosquitoes |
| (KOOV) |
| Wongal virus-MRM168 | mosquitoes |
| (WONV) |
| Madrid virus |
|
|
|
| Madrid virus - Pan BT4075 | mosquitoes |
| (MADV) |
| Madrid virus - BT4075 | mosquitoes | [S: DQ188957] | (MADV) |
| Main Drain virus |
|
|
|
| Main Drain virus-BSF5015 | mosquitoes, culicoid flies | [S: X73469] | (MDV) |
| Manzanilla virus |
|
|
|
| Buttonwillow virus- A7956 | culicoid flies |
| (BUTV) |
| Ingwavuma virus- SA An4165 | mosquitoes |
| (INGV) |
| Inini virus- CaAn1093a | N.D. |
| (INIV) |
| Manzanilla virus- TRVL3587 | N.D. |
| (MANV) |
| Mermet virus- AV-782 | mosquitoes |
| (MERV) |
| Marituba virus |
|
|
|
| Gumbo Limbo virus- FE3-71H | mosquitoes | [S: DQ188953] | (GLV) |
| Marituba virus- BeAn15 | mosquitoes | [M: DQ188966] | (MTBV) |
| Marituba virus - BeAr186247 | mosquitoes | [S: DQ188966 | (MTBV) |
| Marituba virus - 63U-11 | mosquitoes |
| (MTBV) |
| Murutucu virus-BeAn974 | mosquitoes | [S: DQ188947; M: DQ188963] | (MURV) |
| Nepuyo virus- TRVL18462 | mosquitoes | [S: DQ188950] | (NEPV) |
| Restan virus-TRVL51144 | mosquitoes | [S: DQ188956; M: DQ188965] | (RESV) |
| Minatitlan virus |
|
|
|
| Minatitlan virus M67U5 | N.D. |
| (MNTV) |
| Palestina virus - 76V1565 | mosquitoes |
| (PLSV) |
| M’Poko virus |
|
|
|
| M’Poko virus- BA365 | mosquitoes |
| (MPOV) |
| Yaba-1 virus | mosquitoes |
| (Y1V) |
| Nyando virus |
|
|
|
| Nyando virus -MP 401 | mosquitoes |
| (NDV) |
| Eret virus – 147- EthAr147 | mosquitoes |
| (ERETV) |
| Olifantsvlei virus |
|
|
|
| Bobia virus- ArB1569 | mosquitoes |
| (BIAV) |
| Dabakala virus- ArA5937-82 | mosquitoes |
| (DABV) |
| Olifantsvlei virus - SAAr5133 | mosquitoes |
| (OLIV) |
| Oubi virus- ArA3673/81 | mosquitoes |
| (OUBIV) |
| Oriboca virus |
|
|
|
| Itaqui virus- BeAn12797 | mosquitoes | [S: DQ188951; M: DQ188962] | (ITQV) |
| Oriboca virus- BeAn17 | mosquitoes | [S: DQ188946; M:DQ188964] | (ORIV) |
| Oriboca virus- BeH142102 | mosquitoes | [S:DQ188969] | (ORIV) |
| Oropouche virus |
|
|
|
| Facey’s Paddock virus- Aus Ch16129 | N.D. |
| (FPV) |
| Oropouche virus- TRVL9760 | mosquitoes, culicoid flies |
| (OROV) |
| Utinga virus- BeAn84785 | N.D. |
| (UTIV) |
| Utive virus | N.D. |
| (UVV) |
| Patois virus |
|
|
|
| Abras virus-75V1183 | mosquitoes |
| (ABRV) |
| Babahoya virus- 75V2858 | mosquitoes |
| (BABV) |
| Pahayokee virus- FE3-52F | mosquitoes |
| (PAHV) |
| Patois virus - BT4971 | mosquitoes |
| (PATV) |
| Shark River virus- FE4-1R | mosquitoes |
| (SRV) |
| Sathuperi virus |
|
|
|
| Douglas virus- CSIRO150 | culicoid flies |
| (DOUV) |
| Sathuperi virus- IG10310 | mosquitoes, culicoid flies |
| (SATV) |
| Simbu virus |
|
|
|
| Simbu virus - SAAr53 | mosquitoes, culicoid flies |
| (SIMV) |
| Shamonda virus |
|
|
|
| Peaton virus- CSIRO110 | culicoid flies |
| (PEAV) |
| Sango virus- IbAn5077 | mosquitoes, culicoid flies |
| (SANV) |
| Shamonda virus-IbAn5550 | culicoid flies |
| (SHAV) |
| Shuni virus |
|
|
|
| Aino virus- JaNAr28 | mosquitoes, culicoid flies | [S: M22011] | (AINOV) |
| Kaikalur virus- VRC713423-2 | mosquitoes |
| (KAIV) |
| Shuni virus-IbAn10107 | mosquitoes, culicoid flies |
| (SHUV) |
| Tacaiuma virus |
|
|
|
| CoAr 1071 virus | mosquitoes |
| (CA1071V) |
| CoAr 3627 virus | mosquitoes |
| (CA3627V) |
| Tacaiuma virus - BeAn73 | mosquitoes | [S: FJ660416] | (TCMV) |
| Tacaiuma virus - H-32580 | mosquitoes |
| (TCMV) |
| Virgin River - SPAr2317 | mosquitoes |
| (VRV) |
| Virgin River virus – 743-366 | mosquitoes |
| (VRV) |
| Tete virus |
|
|
|
| Bahig virus- EgB90 | ticks |
| (BAHV) |
| Matruh virus- EgAn1047-61 | ticks |
| (MTRV) |
| Tete virus - SAAn 3518 | N.D. | [S: FJ660419] | (TETEV) |
| Tsuruse virus- Mag271580 | N.D. |
| (TSUV) |
| Weldona virus | culicoid flies |
| (WELV) |
| Thimiri virus |
|
|
|
| Thimiri virus- VRC66414 | N.D. |
| (THIV) |
| Timboteua virus |
|
|
|
| Timboteua virus- BeAn116382 | mosquitoes |
| (TBTV) |
| Turlock virus |
|
|
|
| Lednice virus- 6118 | mosquitoes |
| (LEDV) |
| Turlock virus - S 1954-847-32 | mosquitoes |
| (TURV) |
| Umbre virus - IG1412 | mosquitoes |
| (UMBV) |
| Wyeomyia virus |
|
|
|
| Anhembi virus- SPAr2984 | mosquitoes |
| (AMBV) |
| BeAr 328208 virus | mosquitoes |
| (BAV) |
| Macaua virus- BeAr306329 | mosquitoes |
| (MCAV) |
| Sororoca virus- BeAr32149 | mosquitoes |
| (SORO?V) |
| Taiassui virus- BeAr671 | mosquitoes |
| (TAIAV) |
| Wyeomyia virus | mosquitoes |
| (WYOV) |
| Zegla virus |
|
|
|
| Zegla virus- BT5012 | N.D. |
| (ZEGV) |
Species names are in italic script; names of isolates are in roman script. Vector type {}, sequence accession numbers [ ] and assigned abbreviations ( ) are also listed. N.D. = no data.
List of other related viruses which may be members of the genus Orthobunyavirus but have not been approved as species
| Leanyer virus - AusNT16701D | {mosquitoes} | (LEAV) |
| Mojui dos Campos virus BeAn276121 | {N.D.} | (MDCV) |
| Termeil virus - BP8090 | {mosquitoes} | (TERV) |
Genus Hantavirus
Type species Hantaan virus
Distinguishing features
The consensus terminal nt sequences of the L, M and S genomic segments are AUCAUCAUCUG… at the 3′ end and UAGUAGUAUGC… at the 5′ end. Viruses are serologically unrelated to members of other genera. Certain hantaviruses are etiologic agents of hemorrhagic fever with renal syndrome or hantavirus (cardio)pulmonary syndrome (HPS, HCPS). The host range of hantaviruses includes rodents and insectivores, and genetically distinct hantaviruses are usually associated with a single host species. Human infection is incidental to viral maintenance and is almost always a dead end in the infection chain, with the exception of a human-to-human transmission of Andes virus. In contrast to other viruses in the family, hantaviruses are not transmitted by arthropods, and both rodent and human infections are acquired by aerosol exposure to infectious virus in rodent urine, faeces or saliva, and less frequently by rodent bite. Hantaviruses cause no detectable cytopathology in vertebrate cell cultures and cause persistent, non-pathogenic infections of rodents. Hantavirus infections in insectivores are not systematically studied yet.
Electron micrographs of negatively stained particles of isolates of Hantaan virus (left, courtesy of C.S. Schmaljohn) and Tula virus (right, courtesy of S. Butcher and J. Hepojöki). The bars represent 100 nm.
Species demarcation criteria in the genus
Species are usually found in unique ecological niches, i.e. in different primary rodent/insectivore reservoir species. Species exhibit at least 7% difference in aa identity on comparison of the complete glycoprotein precursor and nucleocapsid protein sequences (there are some exceptions presumably caused by historically recent host-switching events). Species show at least four-fold difference in two-way cross neutralization tests. Species do not naturally form reassortants with other species.
List of species in the genus Hantavirus
| Andes virus |
|
|
|
| Andes virus - Chile-9717869 | Oligoryzomys longicaudatus | [L: NC_003468; M: NC_003467; S: NC_003466] | (ANDV) |
| Bermejo virus - Oc22531 | Oligoryzomys chacoensis | [S: AF482713] | (BMJV) |
| Lechiguanas virus - 22819 | Oligoryzomys flavescens | [M: AF028022; S:AF482714] | (LECV) |
| Maciel virus - 13796 | Bolomys obscurus | [S: AF482716] | (MCLV) |
| Oran virus - 22996 | Oligoryzomys longicaudatus | [M: AF028024; S: AF482715] | (ORNV) |
| Pergamino virus - 14403 | Akadon azarae | [S: AF482717] | (PRGV) |
| Bayou virus |
|
|
|
| Bayou virus - Louisiana | Oryzomys palustris | [M: L36930; S: L36929] | (BAYV) |
| Black Creek Canal virus |
|
|
|
| Black Creek Canal virus | Sigmodon hispidus | [M: L39950; S: L39949] | (BCCV) |
| Cano Delgadito virus |
|
|
|
| Cano Delgadito virus - VHV-574 | Sigmodon alstoni | [S: DQ285566] | (CADV) |
| Dobrava virus |
|
|
|
| Dobrava virus - Slovenia, prototype | Apodemus flavicollis
| [M: L33685; S: L41916] | (DOBV)
|
| Dobrava virus - Ano-Poroja/Af9/1999 | Apodemus flavicollis | [L: AJ410615; M: AJ410616: S: AJ410617] | (DOBV) |
| El Moro Canyon virus |
|
|
|
| El Moro Canyon virus - RM-97 | Reithrodontomys megalotis | [M: U26828; S: U11427] | (ELMCV) |
| Hantaan virus |
|
|
|
| Hantaan virus - 76-118 prototype | Apodemus agrarius coreae | [L: X55901; M: M14627; S: M14626] | (HTNV) |
| Isla Vista virus |
|
|
|
| Isla Vista virus - MC-SB-47 | Microtus californicus | [S: U19302] | (ISLAV) |
| Khabarovsk virus |
|
|
|
| Khabarovsk virus- MF43 | Microtus maximowiczii, Microtus fortis | [M: AJ011648; S: U35255] | (KHAV) |
| Laguna Negra virus |
|
|
|
| Laguna Negra virus - 510B | Calomys laucha | [M: AF005728; S: AF005727] | (LANV) |
| Muleshoe virus |
|
|
|
| Muleshoe virus - SH-Tx-339 | Sigmodon hispidus | [S: MHU54575] | (MULV) |
| New York virus |
|
|
|
| New York virus - RI-1 | Peromyscus leucopus | [M: U36801; S: U09488] | (NYV) |
| Prospect Hill virus |
|
|
|
| Bloodland Lake virus - MO46 | Microtus ochrogaster | [S: U19303] | (BLLV) |
| Prospect Hill virus - PH1 | Microtus pennsylvanicus | [M: X55129; S: X55128] | (PHV) |
| Puumala virus |
|
|
|
| Puumala virus - Bashkiria Cg18-20 | Myodes glareolus | [L: M63194; M: M29979; S: M32750] | (PUUV) |
| Puumala virus - Sotkamo, prototype | Myodes glareolus | [L: Z66548; M: X61034; S: X61035] | (PUUV) |
| Rio Mamore virus |
|
|
|
| Rio Mamore virus - OM556 | Oligoryzomys microtis
| [S: U52136] | (RIOMV) |
| Rio Segundo virus |
|
|
|
| Rio Segundo virus | Reithrodontomys mexicanus | [S: U18100] | (RIOS) (SAAC) |
| Saaremaa virus |
|
|
|
| Saaremaa virus - Saaremaa160V prototype | Apodemus agrarius agrarius | [L: AJ410618; M: AJ009774; S: AJ009773] | (SAAV) |
| Saaremaa virus - SK/Aa | Apodemus agrarius agrarius | [M: AY961616; S: AY961615] | (SAAV) |
| Seoul virus |
|
|
|
| Seoul virus - HR80-39 | Rattus norvegicus, Rattus rattus | [L: X56492; M: S47716; S: NC_005236] | (SEOV) |
| Seoul virus - SR-11 virus |
| [M: M34882; S: M34881] | (SEOV) |
| Sin Nombre virus |
|
|
|
| Blue River virus - Indiana | Peromyscus leucopus | [M: AF030551] | (BRV) |
| Blue River virus - Oklahoma | Peromyscus leucopus | [M: AF030552] | (BRV) |
| Monongahela virus | Peromyscus maniculatus | [S: U32591] | (MGLV) |
| Sin Nombre virus - Convict Creek 107 |
| [L: L35008; M: L33474; S: L33683] | (SNV) |
| Sin Nombre virus -NMH10 virus | Peromyscus maniculatus | [L: L37901; M: L25783; S: L25784] | (SNV) |
| Thailand virus |
|
|
|
| Thailand virus - Nakhon Ratchasina/Bi0017/2004 | Bandicota indica | [S: AM397664] | (THAIV) |
| Thailand virus -741 prototype | Bandicota indica | [M: L08756; S: AB186420] | (THAIV) |
| Thottapalayam virus |
|
|
|
| Thottapalayam virus - VCR66412 | Suncus murinus | [L: NC_010707; M: NC_010708; S: NC_010704] | (TPMV) |
| Topografov virus |
|
|
|
| Topografov virus | Lemmus sibiricus | [M: AJ011647; S: AJ011646] | (TOPV) |
| Tula virus |
|
|
|
| Tula virus -Moravia/Ma5302V | Microtus arvalis, M. rossiaemeridionalis | [L: AJ005637; M: Z69993; S: Z69991] | (TULV) |
Species names are in italic script; names of isolates are in roman script. Vector type {}, sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Hantavirus but have not been approved as species
| Altai virus | {Sorex araneus} |
| (ALTV) |
| Amur/Soochong virus | {Apodemus peninsulae} | [L: DQ056292; M: AY675353; S:AY675349] | (ASV) |
| Artybash virus | {Sorex spp.} |
| (ARTV) |
| Araraquara virus | {Bolomys lasiuris} |
| (ARAV) |
| Asama virus | {Urotrichus talpoides} |
| (ASAV) |
| Ash River virus | {Sorex cinereus} |
| (ARRV) |
| Calabazo virus | {Zygodontomys brevicauda} |
|
|
| Camp Riley virus | {Blarina brevicauda} |
| (RPLV) |
| Cao Bang virus | {Anourosorex squamipes} |
| (CBNV) |
| Castelo dos Sonhos virus | {unknown} |
|
|
| Choclo virus | {Oligoryzomys fulvescens} |
|
|
| Da Bie Shan virus | {Niviventer confucianus} | [L: DQ989237; M: AB027115; S: AB027523] | (DBSV) |
| Fox Creek virus | {Sorex palustris} |
| (FXCV) |
| Gou virus | {Rattus rattus} (?) | [M: AB027521; S: AF184988] | (GOUV) |
| Hokkaido virus | {Myodes rufocanus} | [S:AB010730] | (HOKV) |
| Iamonia virus | {Blarina carolinensis} |
| (IAMV) |
| Imjin virus | {Crocidura lasiura} | [L: EF641806; M: EF641798; S: EF641806] | (IMJV) |
| Jemez Springs virus | {Sorex monticolus} |
| (JMSV) |
| Lena River virus | {Sorex caecutiens} |
| (LNAV) |
| Limestone Canyon virus | {Peromyscus boylii} | [M: AF07323; S: AF07322] | (LSCV) |
| Kenkeme virus | {Sorex roboratus} |
| (KENV) |
| Muju virus | {Myodes regulus} | [M: EF198413; S: DQ138133] | (MUJV) |
| Powell Butte virus | {Sorex vagrans} |
| (PWBV) |
| Sangassou virus | {Hylomyscus simus} |
| (SANGV) |
| Seewis virus | {Sorex araneus} |
|
|
| Serang virus | {Rattus tanezumi} |
| (SERV) |
| Tanganya virus | {Crocidura theresae} |
| (TGNV) |
| Tualatin River virus | {Sorex trowbridgii} |
| (TLNV) |
| Vladivostok virus | {Microtus fortis} |
| (VLAV) |
| Yuanjiang virus | {Microtus fortis} |
| (YUJV) |
Genus Nairovirus
Type species Dugbe virus
Distinguishing features
Virions are morphologically similar to other members of the family with very small surface units that appear as a peripheral fringe 7 nm in length (Figure 6). The L RNA segment (12.2 kb) is considerably larger than the L segments of other members of the family. The consensus terminal nt sequences of the L, M and S segments are AGAGUUUCU… at the 3′ end and UCUCAAAGA… at the 5′ end. The S segment does not encode a nonstructural protein. The M segment encodes a single precursor polyprotein that is processed by cotranslational cleavage into precursors to both Gn and Gc. In addition to Gn, posttranslational cleavage of preGn yields also a mucin-rich product and a glycoprotein GP38. Posttranslational cleavage of preGc removes a polypeptide from its C-terminus and yields a mature Gc.
The L protein is predicted to be much larger than those of other members of the family but has yet to be identified. Viruses are serologically unrelated to members of other genera. Most viruses are transmitted by ticks: CCHFV, DUGV and SAKV species are transmitted mainly by ixodid ticks and DGKV, HUGV and QYBV species are transmitted mainly by argasid ticks. Some viruses are transmitted transovarially in arthropods.
Species demarcation criteria in the genus
The paucity of biochemical data dictates that nairovirus species are defined by serological reactivities. There are seven species recognized in the genus Nairovirus.
List of species in the genus Nairovirus
| Crimean-Congo hemorrhagic fever virus |
|
|
|
| Crimean-Congo hemorrhagic fever virus - AP92 | culicoid flies | [S:U04958; M: DQ211625; L: DQ211612] | (CCHFV) |
| Crimean-Congo hemorrhagic fever virus - IbAr10200 | ticks | [S:NC 005302; M: NC 005300; L: NC 005301] | (CCHFV) |
| Hazara virus- PakArJC280 | ticks | [S: M86624; M:DQ813514; L: DQ076419] | (HAZV) |
| Khasan virus- LEIV-776P | ticks |
| (KHAV) |
| Dera Ghazi Khan virus |
|
|
|
| Abu Hammad virus - EgArT1194 | ticks |
| (AHV) |
| Abu Mina virus - EgAn4996 | N.D. |
| (ABMV) |
| Dera Ghazi Khan virus - JD254 | ticks |
| (DGKV) |
| Kao Shuan virus -ArT904 | ticks |
| (KSV) |
| Pathum Thani virus - Ar1753 | ticks |
| (PTHV) |
| Pretoria virus - EgArT3089 | ticks |
| (PREV) |
| Dugbe virus |
|
|
|
| Dugbe virus - IbAr1792 | ticks | [S: AF434164; M: M94133; L: U15018] | (DUGV) |
| Nairobi sheep disease virus | ticks, |
| (NSDV) |
| (Ganjam virus) | culicoid flies, mosquitoes |
|
|
| Hughes virus |
|
|
|
| Farallon virus - USA Ar846 | ticks |
| (FARV) |
| Fraser Point virus | ticks |
| (FPV) |
| Great Saltee virus | ticks |
| (GRSV) |
| Hughes virus | ticks |
| (HUGV) |
| Puffin Island virus | ticks |
| (PIV) |
| Punta Salinas virus - Ar888 | ticks |
| (PSV) |
| Raza virus | ticks |
| (RAZAV) |
| Sapphire II virus | ticks |
| (SAPV) |
| Soldado virus - TRVL52214 | ticks |
| (SOLV) |
| Zirqa virus - A2070-1 | ticks |
| (ZIRV) |
| Qalyub virus |
|
|
|
| Bakel virus - ArD41258 | ticks |
| (BAKV) |
| Bandia virus - Dak IPD/A611 | ticks |
| (BDAV) |
| Omo virus | N.D. |
| (OMOV) |
| Qalyub virus - EgAr37000000 | ticks |
| (QYBV) |
| Sakhalin virus |
|
|
|
| Avalon virus (Paramushir virus) | ticks |
| (AVAV) |
| Clo Mor virus - ScotAr7 | ticks |
| (CMV) |
| Kachemak Bay virus | ticks |
| (KBV) |
| Sakhalin virus - LEIV-71C | ticks |
| (SAKV) |
| Taggert virus - MI 14850 | ticks |
| (TAGV) |
| Tillamook virus | ticks |
| (TILLV) |
| Thiafora virus |
|
|
|
| Erve virus - Brest/AN221 | N.D. |
| (ERVEV) |
| Thiafora virus - AnD11411 | N.D. |
| (TFAV) |
Species names are in italic script; names of isolates are in roman script. Vector type {}, sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Nairovirus but have not been approved as species
None reported.
Genus Phlebovirus
Type species Rift Valley fever virus
Distinguishing features
The surface morphology of phleboviruses is distinct in having small round subunits with a central hole (Figure 7). The consensus terminal nucleotide sequences of the L, M and S segments are UGUGUUUC… at the 3′ end and ACACAAAG… at the 5′ end. The S RNA exhibits an ambisense coding strategy, i.e. it is transcribed by the virion RNA polymerase to a subgenomic virus-complementary sense mRNA that encodes the N protein and, from a full-length antigenome S RNA, to a subgenomic virus-sense mRNA that encodes a nonstructural (NSs) protein. The M segment of viruses in the sandfly fever group but not viruses in the Uukuniemi group has a preglycoprotein coding region that codes for a nonstructural protein(s) (NSm). The Gn and Gc glycoproteins were earlier referred to as G1 and G2 based on apparent size on gel electrophoresis. However, the similar sizes of the G1 and G2 proteins resulted in the different G1:G2 order in the M segments of different viruses. The further adoption of the Gn/Gc nomenclature is strongly encouraged so as to achieve more consistency across the Bunyaviridae family. Phleboviruses are antigenically unrelated to members of other genera, but cross-react serologically among themselves to different degrees. Sandfly fever group viruses are transmitted by phlebotomines, mosquitoes or ceratopogonids of the genus Culicoides; Uukuniemi group viruses are transmitted by ticks.
Species demarcation criteria in the genus
The lack of biochemical data for most phleboviruses dictates that the species are defined by the serological relationships, and are distinguishable by four-fold differences in two-way neutralization tests.
List of species in the genus Phlebovirus
| Bujaru virus |
|
|
|
| Bujaru virus - BeAn 47693 | N.D. |
| (BUJV) |
| Munguba virus - BeAr389707 | phlebotomines |
| (MUNV) |
| Candiru virus |
|
|
|
| Alenquer virus - BeH301101 | N.D. |
| (ALEV) |
| Candiru virus - BeH22511 | N.D. |
| (CDUV) |
| Itaituba virus - BeAn213452 | N.D. |
| (ITAV) |
| Nique virus - Nique-9C | phlebotomines |
| (NIQV) |
| Oriximina virus - BeAr385309 | phlebotomines |
| (ORXV) |
| Turuna virus - BeAr352492 | phlebotomines |
| (TUAV) |
| Chilibre virus |
|
|
|
| Cacao virus - VP-437R | phlebotomines |
| (CACV) |
| Chilibre virus VP-118D | phlebotomines |
| (CHIV) |
| Frijoles virus |
|
|
|
| Frijoles virus VP-161A | phlebotomines |
| (FRIV) |
| Joa virus - BeAr371637 | phlebotomines |
| (JOAV) |
| Punta Toro virus |
|
|
|
| Buenaventura virus -CoAr3319 | phlebotomines |
| (BUEV) |
| Punta Toro virus - PanD4021A | phlebotomines | [M: M11156; S: K02736] | (PTV) |
| Rift Valley fever virus |
|
|
|
| Belterra virus - BeAn356637 | N.D. |
| (BELTV) |
| Icoaraci virus - BeAn24262 | phlebotomines, mosquitoes |
| (ICOV) |
| Rift Valley fever virus | mosquitoes | [L: X56464; M: M11157; S: X53771] | (RVFV) |
| Salehabad virus |
|
|
|
| Arbia virus - ISS.Phl.18 | phlebotomines |
| (ARBV) |
| Salehabad virus - I-81 | phlebotomines |
| (SALV) |
| Sandfly fever Naples virus |
|
|
|
| Karimabad virus - I-58 | phlebotomines |
| (KARV) |
| Massila virus | phlebotomines | [L: EU725773; M: EU725772; S: EU725771] | (MASV) |
| Sandfly fever Naples virus - Sabin | phlebotomines |
| (SFNV) |
| Tehran virus - I-47 | phlebotomines |
| (THEV) |
| Toscana virus - ISS.Phl.3 | phlebotomines | [L: X68414; M: X89628; S: X53794] | (TOSV) |
| Uukuniemi virus |
|
|
|
| EgAN 1825-61 virus | N.D. |
| (EGAV) |
| Fin V 707 virus | N.D. |
| (FINV) |
| Grand Arbaud virus - Argas2 | ticks |
| (GAV) |
| Manawa virus - Argas T461 | ticks |
| (MWAV) |
| Murre virus- | N.D. |
| (MURV) |
| Oceanside virus | ticks |
| (OCV) |
| Ponteves virus - Larves 6 | ticks |
| (PTVV) |
| Precarious Point virus - MI 19334 | ticks |
| (PPV) |
| RML 105355 virus | ticks |
| (RMLV) |
| St. Abbs Head virus | ticks |
| (SAHV) |
| Tunis virus | N.D. |
| (TUNV) |
| Uukuniemi virus - S 23 | ticks | [L: D10759; M: M17417; S: M33551] | (UUKV) |
| Zaliv Terpeniya virus | ticks |
| (ZTV) |
Species names are in italic script; names of isolates are in roman script. Vector type {}, sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Phlebovirus but have not been approved as species
| Aguacate virus - VP-175A | {phlebotomines} |
| (AGUV) |
| Ambe virus - BeAr407981 | {phlebotomines} |
| (AMBEV) |
| Anhanga virus - BeAn46852 | {N.D.} |
| (ANHV) |
| Arboledas virus - CoAr170152 | {phlebotomines} |
| (ADSV) |
| Ariquemes virus - BeAr485678 | {phlebotomines} |
| (ARQV) |
| Armero virus - CoAr171096 | {phlebotomines} |
| (ARMV) |
| Arumowot virus - SudAr1284-64 | {mosquitoes} |
| (AMTV) |
| Caimito virus - VP-488A | {phlebotomines} |
| (CAIV) |
| Chagres virus - JW-10 | {phlebotomines, mosquitoes} |
| (CHGV) |
| Corfou virus - PaAr814 | {phlebotomines} |
| (CFUV) |
| Durania virus - CoAr171162 | {phlebotomines} |
| (DURV) |
| Escharte virus - OBS-6528 | {N.D.} |
| (ESCV) |
| Gabek Forest virus - SudAn754-61 | {N.D.} |
| (GFV) |
| Gordil virus - DakAnBR496d | {N.D.} |
| (GORV) |
| Itaporanga virus | {mosquitoes} |
| (ITPV) |
| Ixcanal virus - CA Ar170897 | {phlebotomines} |
| (IXCV) |
| Jacunda virus - BeAn428329 | {N.D.} |
| (JANV) |
| Leticia virus - CoAr171616 | {phlebotomines} |
| (LTCV) |
| Mariquita virus - Mariquita A | {phlebotomines} |
| (MRQV) |
| Morolillo virus - HTN351 | {N.D.} |
| (MOLV) |
| Morumbi virus - BeH475236 | {N.D.} |
| (MRBV) |
| Mucura virus - BeAr455230 | {mosquitoes} |
| (MRAV) |
| Odrenisrou virus - ArA1131/80 | {mosquitoes} |
| (ODRV) |
| Pacui virus - BeAn27326 | {phlebotomines} |
| (PACV) |
| Rio Grande virus - TBM3-24 | {N.D.} |
| (RGV) |
| Salobo virus - BeAn578142 | {N.D.} |
| (SBOV) |
| Sandfly fever Sicilian virus | {phlebotomines} | [S: J04418] | (SFSV) |
| Saint-Floris virus -DakAnBR512d | {N.D.} |
| (SAFV) |
| Serra Norte virus - BeH505240 | {N.D.} |
| (SRNV) |
| Tapara virus - BeAr413570 | {phlebotomines} |
| (TAPV) |
| Uriurana virus - BeAr479776 | {phlebotomines} |
| (URIV) |
| Urucuri virus - BeAn100049 | {N.D.} |
| (URUV) |
Genus Tospovirus
Type species Tomato spotted wilt virus
Distinguishing features
Morphogenesis occurs in clusters in the cisternae of the endoplasmic reticulum of host cells. Nucleocapsid material may accumulate in the cytoplasm in dense masses; these masses may be composed of defective particles. The morphology of a tospovirus is shown in Figure 8. The consensus terminal sequences of the L, M and S genomic segments are UCUCGUUA… at the 3′ end and AGAGCAAU… at the 5′ end. Both the M and S segment RNAs of tospoviruses utilize an ambisense coding strategy. The virion glycoproteins Gn and Gc are encoded in the complementary-sense RNA of the M segment, and a nonstructural protein, NSm, is encoded in the genome-sense RNA. The S segment encodes the nucleocapsid protein in the complementary-sense RNA and a nonstructural protein, NSs, in the genome-sense RNA. The NSm protein represents the viral (cell-to-cell) movement protein, present in all plant-pathogenic viral taxa and essential for systemic infection of a plant. At the front of infection NSm assembles into tubular structures that penetrate through plasmodesmata thus facilitating nucleocapsids to translocate to the next cell. NSs represents the suppressor of (antiviral) RNAi and can bind both long dsRNA and short dsRNA (i.e. siRNA ans miRNA). In virulent isolates of tospoviruses NSs is highly expressed and may then form paracrystalline or filamentous inclusions in infected plant cells.
At least 13 species of thrips in the genera Frankliniella (9), Thrips (2), Scirtothrips (1) and Ceratothripoides (1) have been reported to transmit tospoviruses, and the Gn and/or Gc glycoproteins are involved in virus–vector interactions. Transmission can also be achieved through infected plant sap. For isolates of the type species Tomato spotted wilt virus, more than 925 plant species belonging to 70 botanical families are known to be susceptible whereas the other tospoviruses have much narrower host ranges.
Species demarcation criteria in the genus
Species are defined on the basis of their vector specificity, their plant host range, serological relationships of the N protein and on the criterion that their N protein sequence should show less than 90% aa identity with that of any other described tospovirus species.
List of species in the genus Tospovirus
| Groundnut bud necrosis virus |
|
|
|
| Groundnut bud necrosis virus (Peanut bud necrosis virus) | {Frankliniella occidentalis, Thrips palmi} | [L: AF025538; M: U42555; S: U27809] | (GBNV) |
| Groundnut ringspot virus |
|
|
|
| Groundnut ringspot virus | {Frankliniella gemina, F. occidentalis, F. schultzei} |
| (GRSV) |
| Groundnut yellow spot virus |
|
|
|
| Groundnut yellow spot virus (Peanut yellow spot virus) | {N.D.} | [S: AF013994] | (GYSV) |
| Impatiens necrotic spot virus |
|
|
|
| Impatiens necrotic spot virus | {Frankliniella occidentalis} | [L: X93218; M: M74904; S: X66972] | (INSV) |
| Tomato chlorotic spot virus |
|
|
|
| Tomato chlorotic spot virus | {Frankliniella occidentalis, F. schultzei, F. intonsa} | [S(N) :S54325] | (TCSV) |
| Tomato spotted wilt virus |
|
|
|
| Tomato spotted wilt virus | {Frankliniella bispinosa, F. cephalica, F. gemina, F. fusca, F. intonsa, F. occidentalis, F. schultzei, F. setosus, Thrips tabaci} | [L: (BR-01) D10066; M: (BR-01) S48091; S:(BR-01) D00645; S: (B) L12048;S: (BL) L20953;S: (L3) D13926] | (TSWV) |
| Watermelon silver mottle virus |
|
|
|
| Watermelon silver mottle virus | {Thrips palmi} | [M: U75379;S: Z46419] | (WSMoV) |
| Zucchini lethal chlorosis virus |
|
|
|
| Zucchini lethal chlorosis virus | {Frankliniella zucchini} | [S(N): AF067069] | (ZLCV) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Vector type {}, sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Tospovirus but have not been approved as species
| Alstromeria necrotic streak virus | {Frankliniella occidentalis} | [S: GQ478668 (N)] | (ANSV) |
| Calla lily chlorotic spot virus | {N.D.} | [L: FJ822961; M: FJ822962] | (CCSV) |
| Capsicum chlorosis virus(Gloxinia tospovirus) | {Ceratotripoides claratris} | [L: DQ256124, M: DQ256125;S: DQ256123] | (CaCV) |
| (Thailand tomato tospovirus) |
|
|
|
| Chrysanthemum stem necrosis virus | {Frankliniella occidentalis} | [S(N): AF067068] | (CSNV) |
| Groundnut chlorotic fan-spot virus | {Scirtothrips dorsalis} | [ S(N):AF080526] | (GCFSV) |
| Iris yellow spot virus | {N.D.} | [L: FJ623474; M: AF214014; S: AF001387] | (IYSV) |
| Melon severe mosaic virus | {N.D.} | [S: EU275149] | (MSMV) |
| Melon yellow spot virus | {Thrips palmi} | [L: AB061774; M: AB061773; S: AB038343] | (MYSV) |
| Physalis severe mottle virus | {N.D.} | [S: AF067151] | (PhySMV) |
| Polygonum ringspot virus | {N.D.} | [M: EU271753; S: EF445397] | (PolRSV) |
| Tomato necrosis virus | {N.D.} | [M: AY647437] | (TNeV) |
| Tomato necrotic ringspot virus | {N.D.} | [M: FJ947152; S: FJ489600] | (TNRV) |
| Tomato yellow (fruit) ring virus | {Thrips tabaci} | [S: AY686718] | (TYRV) |
| Tomato zonate spot virus | {N.D.} | [L: EF552435; M: EF552434; S: EF552433] | (TZSV) |
| Watermelon bud necrosis virus | {Thrips palmi} | [M: FJ694963; S: EU249351] | (WBNV) |
List of other related viruses which may be members of the family Bunyaviridae but have not been approved as species
There are seven groups (19 viruses) and 21 ungrouped viruses which have not been assigned to a recognized genus in the family Bunyaviridae. For most, no biochemical characterization of the viruses has been reported to determine their taxonomic status.
| Grouped viruses: |
|
| Bhanja virus - IG690 | (BHAV) |
| Forecariah virus - ArK4927 | (FORV) |
| Kismayo virus - A3641 | (KISV) |
| Kaisodi virus - IG14132 | (KSOV) |
| Lanjan virus - Mal TP94 | (LJNV) |
| Silverwater virus - Can131 | (SILV) |
| Mapputta virus - AusNRM186 | (MAPV) |
| Gan Gan virus - NB6057 | (GGV) |
| Maprik virus - MK7532 | (MPKV) |
| Trubanaman virus - AusMRM3630 | (TRUV) |
| Okola virus - Dak YM50/64 | (OKOV) |
| Tanga virus -TanzMP1329 | (TANV) |
| Resistencia virus - AG80-504 | (RTAV) |
| Antequera virus - AG80-226 | (ANTV) |
| Barranqueras virus -AG80-381 | (BQSV) |
| Yogue virus - DakAnD5634 | (YOGV) |
| Kasokero virus - UGZ52969 | (KASV) |
| Ungrouped viruses: |
|
| Bangui virus - DakHB745 | (BGIV) |
| Belem virus - BeAn141106 | (BLMV) |
| Belmont virus - R8659 | (BELV) |
| Bobaya virus - AnB2208d | (BOBV) |
| Caddo Canyon virus | (CDCV) |
| Chim virus - LEIV-858Uz | (CHIMV) |
| Enseada virus - 75V25880 | (ENSV) |
| Issyk-Kul virus - LEIV-315K | (ISKV) |
| Keterah virus - P6-1361 | (KTRV) |
| Kowanyama virus - Aus MRM1178 | (KOWV) |
| Lone Star virus - USA TMA1381 | (LSV) |
| Pacora virus - PanJ19 | (PCAV) |
| Para virus - BeAn280577 |
|
| Razdan virus - LEIV-2741Ar | (RAZV) |
| Santarem virus - BeAn238758 | (STMV) |
| Sunday Canyon virus - RML52301-11 | (SCAV) |
| Tai virus - ArA 94/79 | (TAIV) |
| Tamdy virus - LEIV-1308z | (TDYV) |
| Tataguine virus - DaKIPD/A252 | (TATV) |
| Wanowrie virus - IG700 | (WANV) |
| Witwatersrand virus - SAAr1062 | (WITV) |
| Yacaaba virus - NB6028 | (YACV) |
Phylogenetic relationships within the family
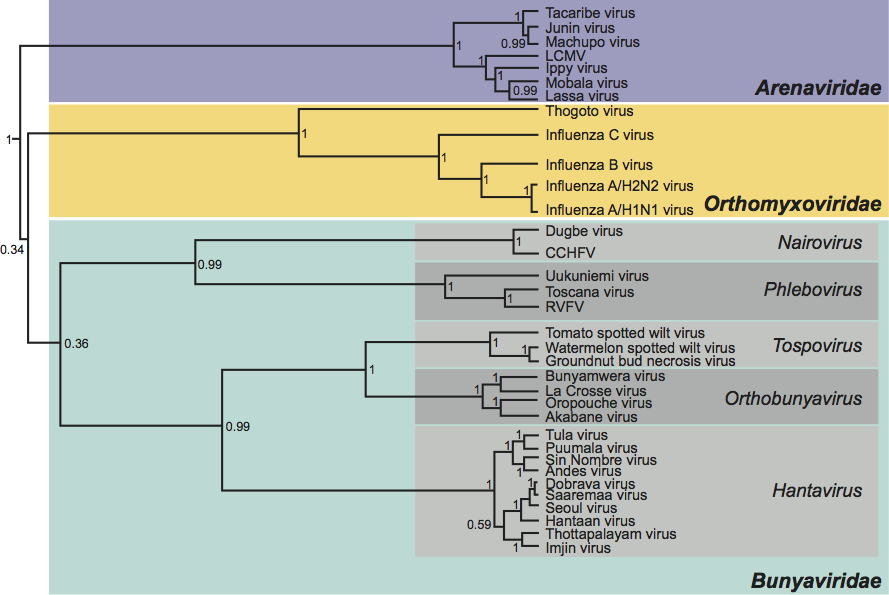
As documented above, the analogous genes and gene products of viruses in the different genera vary widely in size, and there is little obvious global similarity at either nt or aa level. Attempts to produce convincing alignments of either genome segments or structural proteins from which to generate phylogenetic trees have so far proved unsuccessful, with the exception of the putative polymerase domain of the L proteins. Such analysis suggests that viruses in the family Bunyaviridae fall into two major lineages, comprising orthobunyaviruses, hantaviruses and tospoviruses on one, and nairoviruses and phleboviruses in the other. The significant point to note is that L protein phylogeny does not segregate with the use of an ambisense coding strategy. (See Figure 9.)
Similarity with other taxa
The plant-infecting tenuiviruses show some similarities to members of the family Bunyaviridae, particularly the genus Phlebovirus. Tenuiviruses have a ssRNA genome comprising four or five segments that encode proteins using a negative or ambisense coding strategy. The tenuivirus RNA terminal sequences are conserved and the 3′ and 5′-sequences exhibit inverted complementarity; the conserved 3′-sequence, UGUGUUUCAG…, is similar to the consensus phlebovirus sequence. Tenuiviruses employ a cap-snatching mechanism to prime viral mRNA synthesis, similar to members of the family Bunyaviridae. Weak sequence homology has been noted between the Rice stripe virus 94 kDa protein and phlebovirus glycoproteins, and between tenuivirus nucleocapsid proteins and those of phleboviruses.
Derivation of names
Bunya: from Bunyamwera, place in Uganda, where type virus was isolated.
Hanta: from Hantaan, river in South Korea near where type virus was isolated.
Nairo: from Nairobi sheep disease, first reported disease caused by member virus.
Phlebo: refers to phlebotomine vectors of sandfly fever group viruses; Greek phlebos, “vein”.
Tospo: from tomato spotted wilt virus.
Further reading
Elliott, 1996 R.M. Elliott, The Bunyaviridae. In: R.M. Elliott, The Bunyaviridae. Plenum Press, New York1996.
Goldbach and Kuo, 1996 R. Goldbach, G. Kuo, Introduction (Tospoviruses and Thrips). Acta Horticult. 431 (1996) 21–26.
Karabatsos, 1985 N. Karabatsos, International Catalogue of Arboviruses Including Certain Other Viruses of Vertebrates. In: N. Karabatsos, International Catalogue of Arboviruses Including Certain Other Viruses of Vertebrates. American Society of Tropical Medicine and Hygiene, San Antonio, Texas1985.
Plyusnin, 2002 A. Plyusnin, Genetics of hantaviruses: implications to taxonomy. Arch. Virol. 147 (2002) 665–682.
Schmaljohn and Nichol, 2007 C.S. Schmaljohn, S. Nichol, D.M. Knipe, P. Howley, BunyaviridaeFields Virology. In: D.M. Knipe, P. Howley, Fields Virology. Lippincott, Williams and Wilkins, Philadelphia20071741–1789.
Schmaljohn and Nichol, 2001 Schmaljohn, C.S. and Nichol, S.T. (Eds.) (2001). Hantaviruses. Current Topics in Microbiology and Immunology 256. Berlin: Springer Verlag.
Contributed by
Plyusnin, A., Beaty, B.J., Elliott, R.M., Goldbach, R., Kormelink, R., Lundkvist, Å., Schmaljohn, C.S. and Tesh, R.B
Figures
Figure 1 (Left) Diagrammatic representation of an orthobunyavirus virion in cross-section. The surface spikes comprise two glycoproteins termed Gn and Gc (previously referred to as G1 and G2). The three helical nucleocapsids are circular and comprise one each of the unique ssRNA segments (L, large; M, medium; S, small) encapsidated by N protein and associated with the L protein
(courtesy of R. Pettersson).
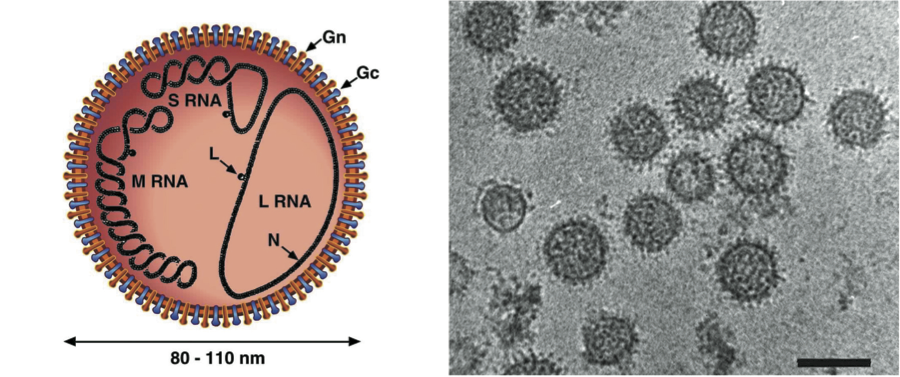
Figure 2 Coding strategies of genome segments of members of the family Bunyaviridae.
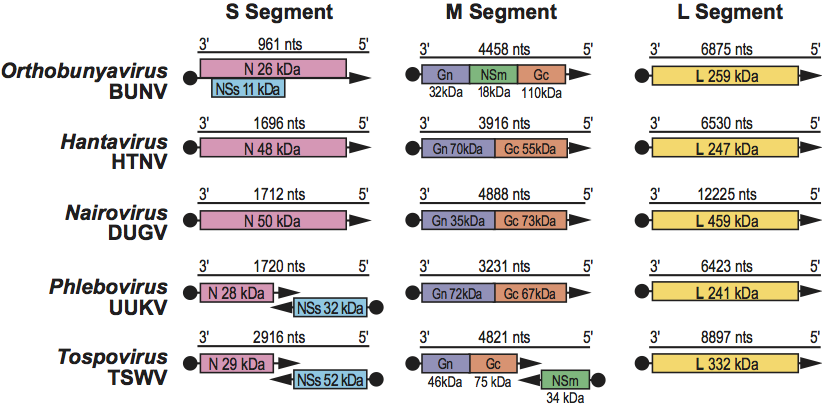
Figure 3 Transcription and replication scheme of genome segments of members of the family Bunyaviridae for a negative-strand segment (left) and for an ambisense segment (right). The genome RNA and the positive sense viral complementary RNA, known as anti-genome RNA, are only found as ribonucleoprotein complexes and are encapsidated by N protein. The mRNA species contain host-derived primer sequences at their 5 ends () and are truncated at the 3 end relative to the vRNA template; the mRNAs are not polyadenylated. For the ambisense TSWV M and S RNA-derived subgenomic transcripts, termination occurs in the intergenic region, likely due to folding of a predicted AU-rich hairpin structure in nascent transcripts.
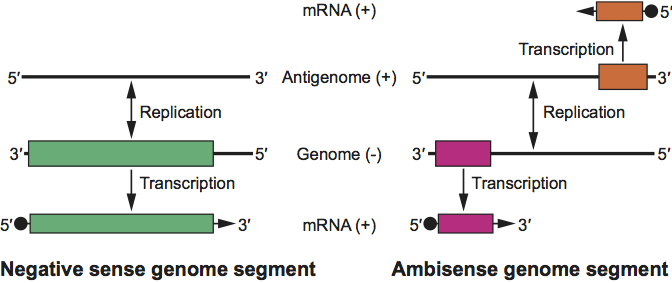
Figure 4 Electron micrograph of negatively stained particles of California encephalitis virus strain La Crosse virus. The bar represents 100 nm.
(Courtesy of D. H. L. Bishop.)
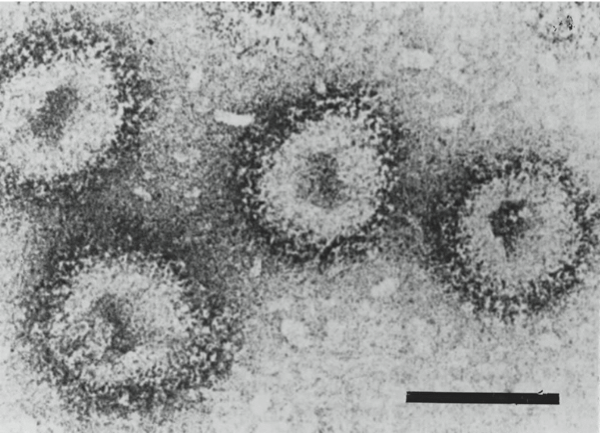
Figure 5 Electron micrographs of negatively stained particles of isolates of Hantaan virus (left, courtesy of C.S. Schmaljohn) and Tula virus (right, courtesy of S. Butcher and J. Hepojki). The bars represent 100 nm.
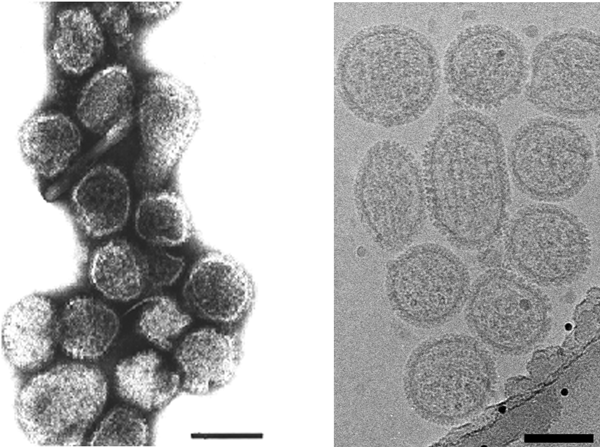
Figure 6 Electron micrograph of negatively stained particles of Crimean-Congo hemorrhagic fever virus (CCHFV). The bar represents 100 nm.
(Courtesy of C. S. Schmaljohn.)
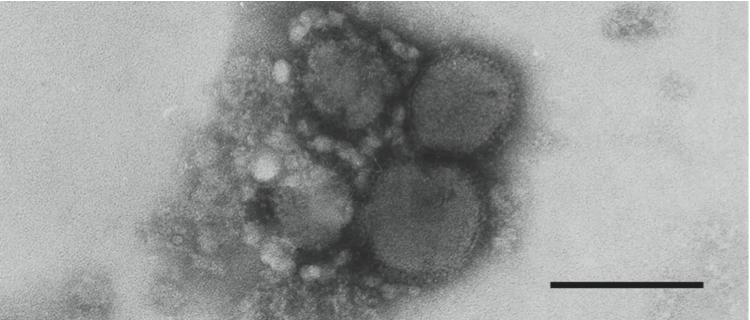
Figure 7 (Left) Electron micrograph of negatively stained particles of Rift Valley fever virus. The bar represents 50 nm
(courtesy of A. Freiburg and R. Flick).
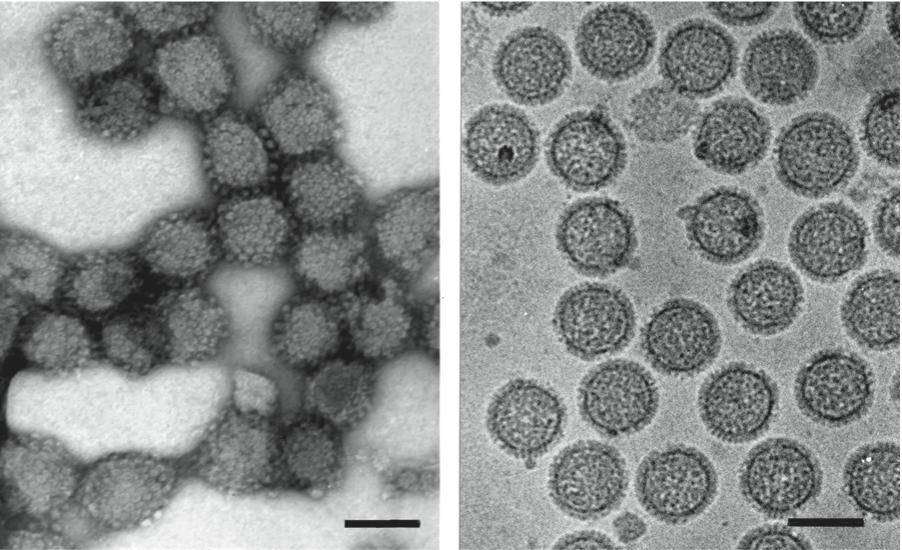
Figure 8 Electron micrograph of negatively stained particles of Tomato spotted wilt virus (TSWV). The bar represents 100 nm.
(Courtesy of Dr Jan van Lent.)
Figure 9 Phylogenetic tree of aligned core polymerase domains from the L proteins of members of the family Bunyaviridae and from analogous proteins of other segmented (Arenaviridae, Orthomyxoviridae) negative strand RNA viruses. The tree was reconstructed using the Bayesian Monte Carlo Markov Chain method in BEAST (http://beast.bio.ed.ac.uk/). A maximum clade credibility tree is shown with mean branch lengths (substitutions per site), and Bayesian posterior probabilities given at the nodes.
(Courtesy of T. Sironen.)