Family: Polymycoviridae
Ioly Kotta-Loizou and Robert H. A. Coutts
The citation for this ICTV Report chapter is the summary published as Kotta-Loizou et al.,(2022):
ICTV Virus Taxonomy Profile: Polymycoviridae 2022, Journal of General Virology, 103:001747
Corresponding author: Ioly Kotta-Loizou ([email protected])
Edited by: Sead Sabanadzovic and Peter Simmonds
Posted: March 2022, updated May 2024
Summary
Polymycoviridae is a family of small viruses with multi-segmented and non-conventionally encapsidated double-stranded (ds) RNA genomes (Table 1 Polymycoviridae). Typically, polymycoviruses have four genomic segments, although some have up to eight. The dsRNA segments together comprise a genome of 7.5–12.5 kbp. Polymycoviruses infect fungi (ascomycetes and basidiomycetes) and oomycetes. The one genus of the family, Polymycovirus, includes 10 species.
Table 1 Polymycoviridae. Characteristics of members of the family Polymycoviridae.
| Characteristic | Description |
| Example | Aspergillus fumigatus tetramycovirus 1 (dsRNA1: HG975302; dsRNA2: HG975303; dsRNA3: HG975304; dsRNA4: HG975305), species Polymycovirus aspergilli, genus Polymycovirus |
| Virion | Non-conventionally encapsidated dsRNA, coated with viral protein |
| Genome | A total of 7.5–12.5 kbp of dsRNA in a multipartite genome (usually four segments, up to eight) |
| Replication | Both dsRNA and ssRNA can be isolated from infected fungal hosts. Virions accumulate in the cytoplasm |
| Translation | From positive-sense transcripts of genomic dsRNAs |
| Host range | Fungi (ascomycetes and basidiomycetes), oomycetes |
| Taxonomy | Realm Riboviria; one genus Polymycovirus including 10 species |
Virion
Morphology
The majority of known polymycoviruses form non-conventional virions, whereby genomic dsRNA is coated by a viral protein (Kanhayuwa et al., 2015, Zhai et al., 2016, Kotta-Loizou and Coutts 2017, Niu et al., 2018). Only one polymycovirus, the eight-segmented Colletotrichum camelliae filamentous virus 1 (CcFV1), is believed to have a filamentous capsid (Jia et al., 2017), ranging from 10 to 12 nm in width and over 1000 nm in length (Figure 1 Polymycoviridae).
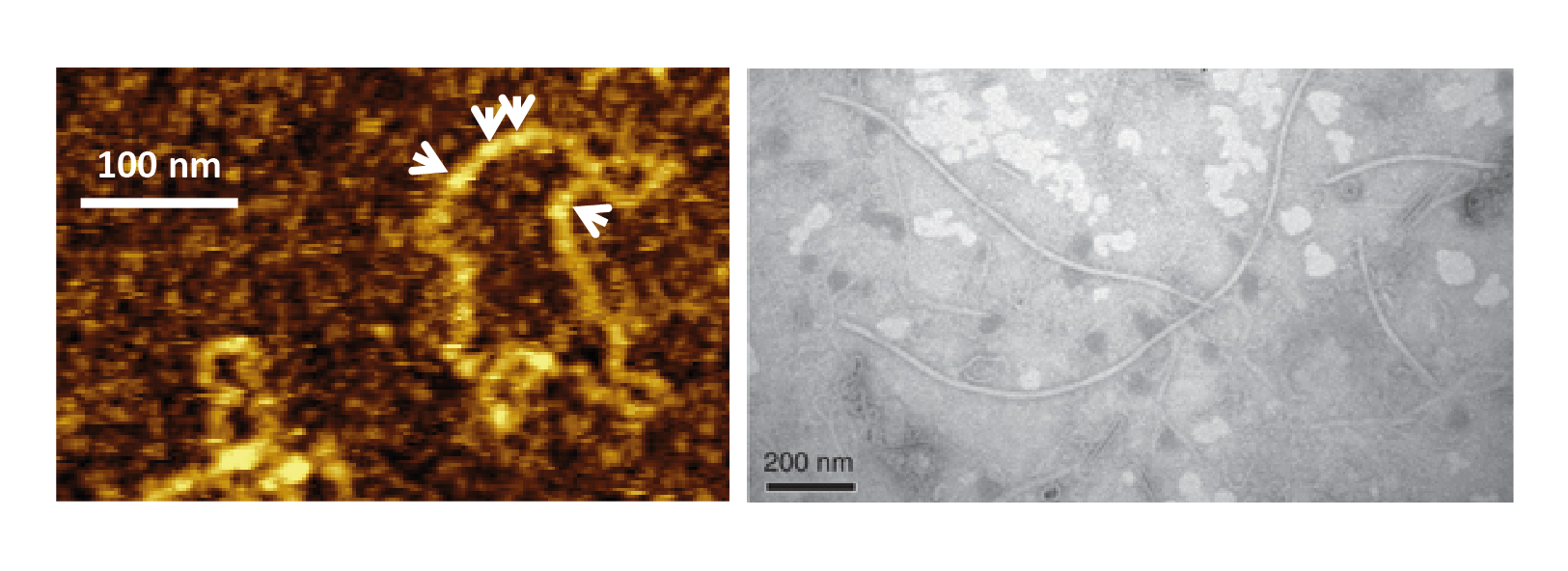 |
| Figure 1 Polymycoviridae. Polymycovirus virions. (Left) Visualisation of purified Aspergillus fumigatus tetramycovirus 1 non-conventional virions using atomic force microscopy (AFM); white arrows indicate the viral protein coating the dsRNA genome (adapted from (Kanhayuwa et al., 2015)). (Right) Visualisation of purified Colletotrichum camelliae filamentous virus 1 conventional virions by transmission electron microscopy (TEM) (adapted from (Jia et al., 2017) under http://creativecommons.org/licenses/by/4.0/). |
Physicochemical and physical properties
Following centrifugation in stepwise sucrose gradients (60–10% with 10% sucrose increments, or 40–15% with 5% sucrose increments), CcFV-1 conventional, filamentous virions are found in the gradient zone from 20 to 30% sucrose (Jia et al., 2017). Information is not available for other members of the family forming non-conventional virions.
Nucleic acid
Virions contain from four up to eight (Jia et al., 2017, Mahillon et al., 2019) linear, dsRNA segments, ranging from 0.8 to 2.5 kbp.
Proteins
The dsRNAs are coated by the intrinsically disordered proline-alanine-serine rich protein (PASrp; 18–32 kDa) (Kanhayuwa et al., 2015, Zhai et al., 2016, Kotta-Loizou and Coutts 2017, Niu et al., 2018). In the case of CcFV-1, PASrp is the single major polypeptide forming the filamentous capsids (Jia et al., 2017), although it is possible that the ability to form a capsid is dependent on the presence of additional viral proteins.
Genome organization and replication
Polymycoviruses have multipartite genomes comprising four to eight linear dsRNA segments; each segment is monocistronic, with an open reading frame (ORF) flanked by non-coding regions (NCRs). The terminal sequences of the 5′- and 3′-NCRs are highly conserved among the four-dsRNA segments (Figure 2 Polymycoviridae).
The ORF of dsRNA 1 encodes the RNA dependent RNA polymerase (RdRP) responsible for the replication of the virus. The RdRP of polymycoviruses belongs to the protein family RdRP_1 (Pfam PF00680) and has three conserved motifs (MSDITKWDANMC, YGCMPSGSFYTSLVNTTGNNL and YGDNQLFSEHL) typical of RdRPs in its catalytic domain (Figure 2 Polymycoviridae). The ORF of dsRNA 2 encodes a protein of unknown function, hypothesized to act as a scaffold protein for replication and virion assembly (Kotta-Loizou and Coutts 2017), which contains a conserved N-terminus and a cysteine-rich, zinc finger-like motif (Figure 2 Polymycoviridae), and is rich in arginine repeats (R-R, R-X-R), associated with endoplasmic reticulum (ER) retention signals. The ORF of dsRNA 3 encodes a methyl transferase, responsible for adding a capping structure at the 5′-termini of the positive-sense strands of the viral dsRNAs (Kanhayuwa et al., 2015, Kotta-Loizou and Coutts 2017). Similar to other redox enzymes from all kingdoms of life, the polymycovirus methyl transferases are two-domain proteins (Figure 2 Polymycoviridae), containing a methyltransferase catalytic motif and an N-terminal Rossmann-fold domain. The ORF of dsRNA 4 encodes PASrp. When present, dsRNAs 5–8 encode proteins of unknown function that are non-homologous between different viruses.
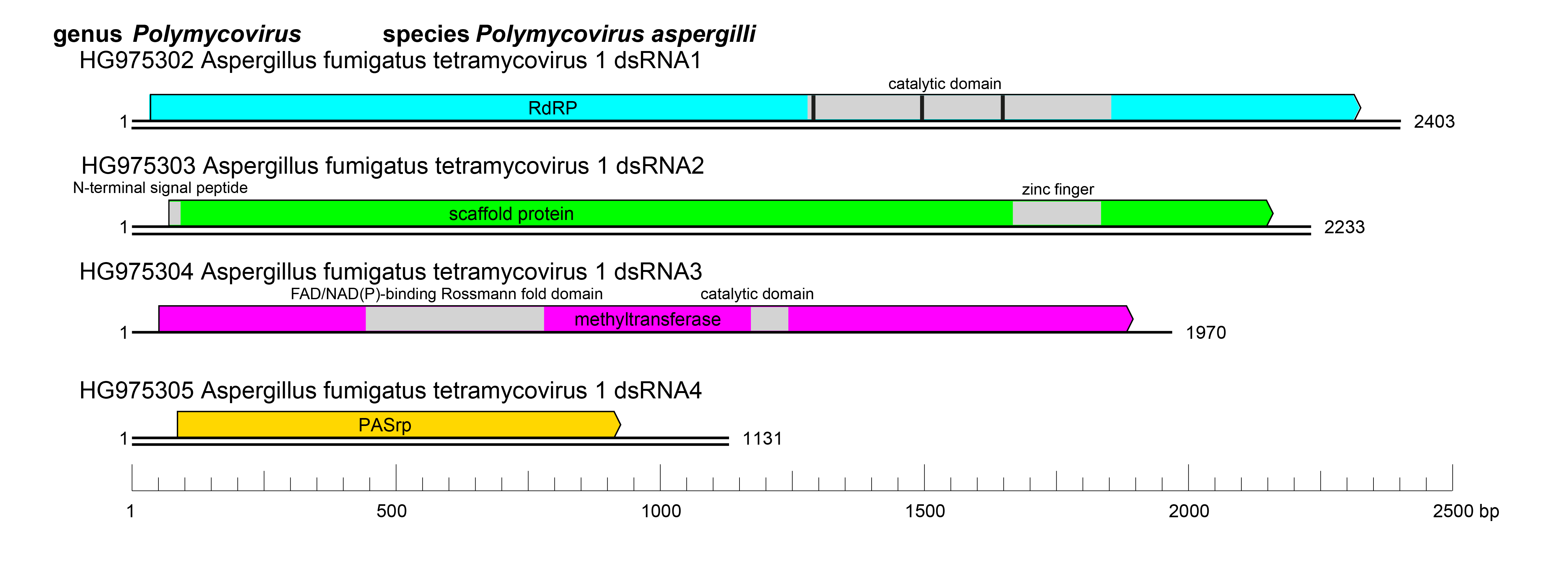 |
| Figure 2 Polymycoviridae. Genome organization of Aspergillus fumigatus tetramycovirus 1 (AfuTmV-1). The genome consists of four monocistronic dsRNA segments. The proteins encoded are represented by rectangular boxes. The grey boxes in dsRNAs 1–3 represent the approximate location of known and/or predicted motifs and domains, such as the RdRP_1 domain (PF00680) in dsRNA1 (with three conserved motifs represented by black bars) and Methyltransf_25 (PF13649) in dsRNA3. |
Biology
Polymycoviruses lack an extracellular phase in their life cycle; they are transmitted intracellularly during cell division and sporogenesis (vertical transmission) and by cell fusion following hyphal anastomosis between compatible fungal strains (horizontal transmission) (Zhai et al., 2016, Filippou et al., 2018). Virions accumulate in the cytoplasm. Polymycoviruses were the first dsRNA viruses found to be infectious not only as purified entities but also as naked dsRNA (Kanhayuwa et al., 2015, Jia et al., 2017, Niu et al., 2018). Polymycovirus infection has been associated with various morphological alterations in the host cells, including changes in pigmentation (Kanhayuwa et al., 2015, Jia et al., 2017, Kotta-Loizou and Coutts 2017, Filippou et al., 2021) and sectoring (Kanhayuwa et al., 2015, Zhai et al., 2016, Kotta-Loizou and Coutts 2017). Polymycovirus infection has also been associated with decreased host growth (Zhai et al., 2016, Jia et al., 2017) and host virulence against plants (Zhai et al., 2016, Jia et al., 2017), insects (Pitaluga et al., 2020) and mammals (Takahashi-Nakaguchi et al., 2020), or increased sporulation (Filippou et al., 2021), host growth (Kotta-Loizou and Coutts 2017, Filippou et al., 2021) and host virulence in the greater wax moth (Galleria mellonella) infection model (Özkan and Coutts 2015, Kotta-Loizou and Coutts 2017). Increased host sensitivity to antifungals (Niu et al., 2018) and to the bacterium Pseudomonas aeruginosa (Nazik et al., 2021) has also been reported. Although the molecular mechanisms underpinning the above phenotypes have not been elucidated, there is evidence that polymycoviruses modulate host carbon, nitrogen and iron metabolism (Filippou et al., 2021, Patil et al., 2021). Finally, polymycoviruses are targeted by the host RNA silencing machinery (Özkan et al., 2017).
Antigenicity
Polyclonal antibodies with high affinity and specificity are produced in mice and rabbits immunized with purified or recombinant PASrp (Zhai et al., 2016, Jia et al., 2017).
Derivation of names
Polymycoviridae, Polymycovirus: poly meaning “many” in Greek; indicating the numerous genomic dsRNA segments of the virus and myco meaning “fungus” in Greek.
Genus demarcation criteria
There is only one genus Polymycovirus in the family Polymycoviridae.
Relationships within the family
There is only one genus Polymycovirus in the family Polymycoviridae accommodating all species (Figure 3 Polymycoviridae).
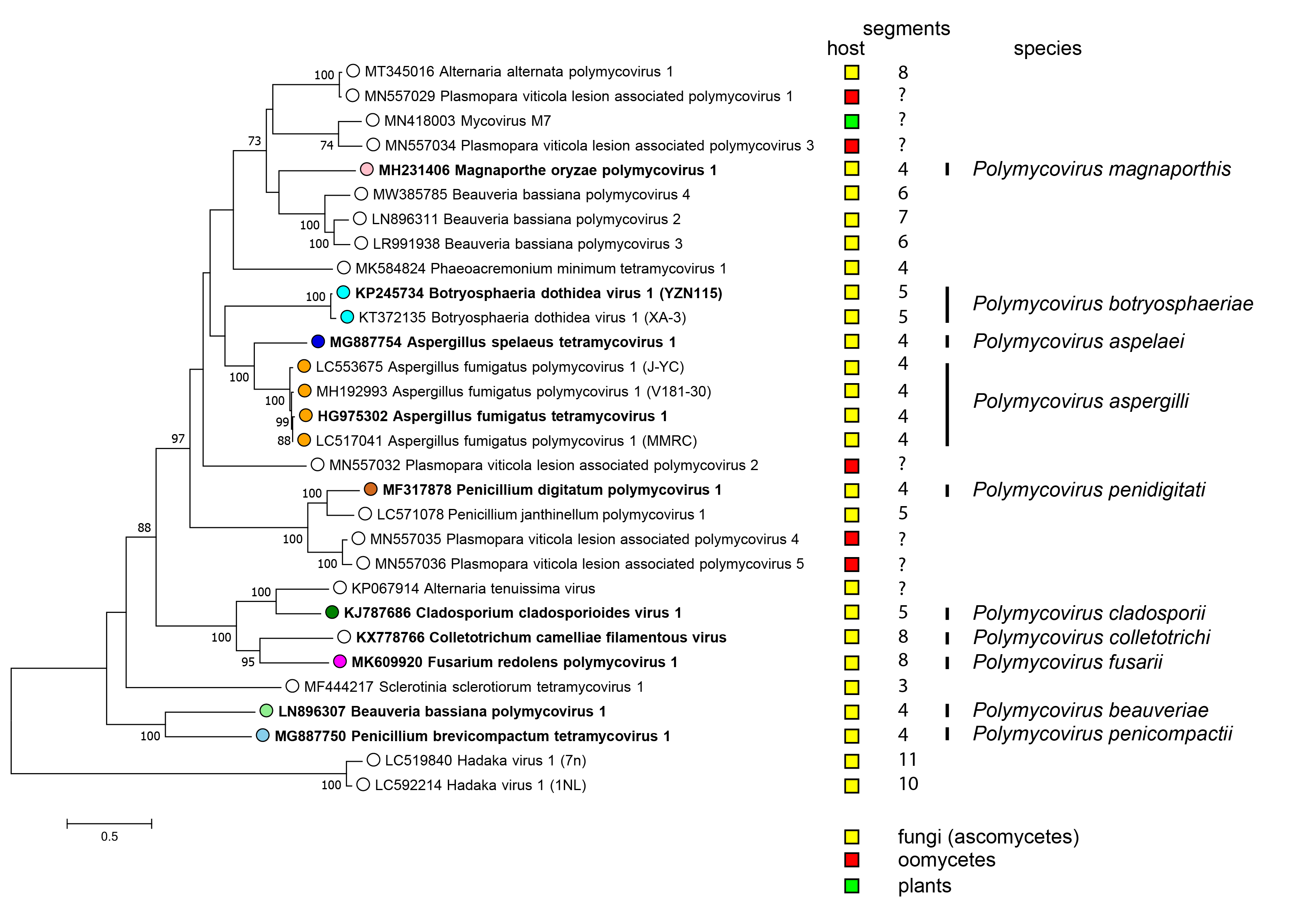 |
| Figure 3 Polymycoviridae. Phylogenetic analysis of members of the family Polymycoviridae and related viruses based on amino acid sequences of their RdRP. A multiple alignment of RdRP amino acid sequences was produced using MUSCLE (Edgar 2004), the LG+G+I+F substitution model was used and a maximum likelihood phylogenetic tree was created by the program MEGA X (Kumar et al., 2018). Bootstrap percentages (100 replicates) are shown where over 70%. Tips labelled with open circles indicate currently unclassified viruses. Exemplar isolates of each species are indicated by bold text. Virus hosts or associations are indicated by squares coloured yellow (fungi - ascomycetes), red (oomycetes) and green (plants). The number of dsRNA genomic segments is indicated, if known. |
Relationships with other taxa
Polymycoviridae belongs to the realm Riboviria, accommodating RNA viruses; however, the family’s classification within Riboviria and its relationships with other RNA virus families has yet to be elucidated. Viruses in the family Polymycoviridae are most closely related to Hadaka virus 1 isolates, positive-sense (+) single-stranded (ss) RNA viruses in the family Hadakaviridae (Sato et al., 2020). The RdRPs of polymycoviruses are also related to those of (+)ssRNA viruses in the families Astroviridae, Caliciviridaeand Picornaviridae, and of the dsRNA family Partitiviridae in the phylum Pisuviricota. The GDNQ motif, typically found in negative-sense ssRNA viruses of the order Monogenavirales, is conserved in the RdRPs encoded by all members of Polymycoviridae, instead of the GDD motif found in most dsRNA and positive-sense ssRNA viruses. Polymycoviruses appear to be intermediate between dsRNA and (+)ssRNA viruses, as well as between encapsidated and capsidless RNA viruses (Kanhayuwa et al., 2015), since members of the family may or may not form a conventional virions.
Related, unclassified viruses
| Virus | Accession number |
| Alternaria alternata polymycovirus 1 | dsRNA 1: MT345016; dsRNA 2: MT345017; dsRNA 3: MT345018; dsRNA 4: MT345019; dsRNA 5: MT345020; dsRNA 6: MT345021; dsRNA 7: MT345022; dsRNA 8: MT345023 |
| Alternaria tenuissima virus | dsRNA 1: KP067914 |
| Alternaria sp. FA0703 sequence | dsRNA 2: FJ595830; dsRNA 5: FJ595831; dsRNA 6: FJ595830 |
| Beauveria bassiana polymycovirus 2 | dsRNA 1: LN896311; dsRNA 6: LN896312; dsRNA 7: LN896313 |
| Beauveria bassiana polymycovirus 3 | dsRNA 1: LR991938; dsRNA 2: LR991939; dsRNA 3: LR991940; dsRNA 4: LR991941; dsRNA 5: LR991942; dsRNA 6: LR991943 |
| Beauveria bassiana polymycovirus 4 | dsRNA 1: MW385785; dsRNA 2: MW385786; dsRNA 3: MW385787; dsRNA 4: MW385788; dsRNA 5: MW385789; dsRNA 6: MW385790 |
| Melampsora lini sequences | dsRNA 3: X64371 |
| Mycovirus M7 | dsRNA 1: MN418003 |
| Phaeoacremonium minimum tetramycovirus 1 | dsRNA 1: MK584824; dsRNA 2: MK584825; dsRNA 3: MK584826;dsRNA 4: MK584827 |
| Penicillium janthinellum polymycovirus 1 | dsRNA 1: LC571078; dsRNA 2: LC571079; dsRNA 3: LC571080; dsRNA 4: LC571081; dsRNA 5: LC571082 |
| Plasmopara viticola lesion associated polymycovirus 1 | dsRNA 1: MN557029; dsRNA 3: MN557030 |
| Plasmopara viticola lesion associated polymycovirus 2 | dsRNA 1: MN557032; dsRNA 2: MN557031; dsRNA 4: MN557033 |
| Plasmopara viticola lesion associated polymycovirus 3 | dsRNA 1: MN557034 |
| Plasmopara viticola lesion associated polymycovirus 4 | dsRNA 1: MN557035 |
| Plasmopara viticola lesion associated polymycovirus 5 | dsRNA 1: MN557036; dsRNA 2: MN557037; dsRNA 4: MN557038 |
| Sclerotinia sclerotiorum tetramycovirus 1 | dsRNA 1: MF444217; dsRNA 2: MF444218; dsRNA 3: MF444219 |
Virus names and virus abbreviations are not official ICTV designations.
Most of these viruses have partially sequenced genomes and the number of genomic segments for each virus is often unknown. Others, such as Alternaria alternata polymycovirus 1, Beauveria bassiana polymycovirus 3 and 4, and Penicillium janthinellum polymycovirus 1 have been completely sequenced and will be included in the next taxonomic update of Polymycoviridae; some of them are likely to represent new species in the family.

