Family: Pseudoviridae
Carlos Llorens, Beatriz Soriano and Mart Krupovic
The citation for this ICTV Report chapter is the summary published as Llorens et al., (2021):
ICTV Virus Taxonomy Profile: Pseduoviridae, Journal of General Virology, 102 (3): 001563
Corresponding author: Carlos Llorens [email protected]) and Mart Krupovic ([email protected])
Edited by: Balázs Harrach and Stuart G. Siddell
Posted: January 2021
PDF: ICTV_Pseudoviridae.pdf
Summary
Pseudoviridae is a family of reverse-transcribing viruses with long terminal repeats (LTRs) belonging to the order Ortervirales. Pseudoviruses are broadly known as Ty1/Copia LTR retrotransposons, which are commonly found integrated in the genomes of a wide range of plants, fungi and animals. Inside the host cell, they form icosahedral virus particles, but unlike most other viruses, do not have an extracellular phase. Members of the family Pseudoviridae share evolutionary history as well as structural and functional features with members of all other families of the order Ortevirales – particularly the Metaviridae and Belpaoviridae – but differ from them in the domain organization of the pol region. The International Committee on Taxonomy of Viruses (ICTV) currently recognizes three genera within this family – Pseudovirus, Hemivirus and Sirevirus – mainly based on the differences in features such as the primer binding site (for pseudoviruses and hemiviruses) or presence of a third env gene (in sireviruses). This report will focus on the three currently recognized genera; a number of related viruses have yet to be formally classified, and the classification of pseudovirids is in need of revision.
Table 1. Pseudoviridae. Characteristics of members of the family Pseudoviridae
| Characteristic | Description |
| Example | Saccharomyces cerevisiae Ty1 virus (M18706), species Pseudovirus unisaccharomycetis |
| Virion | Virions are icosahedral (T=3 or 4) and might be enveloped |
| Genome | Two identical copies of linear single-stranded RNA |
| Replication | Replication by reverse-transcription primed with a host-encoded tRNA |
| Translation | Genomic RNA is translated into one or more polyproteins |
| Host range | Fungi, plants and animals |
| Taxonomy | Realm Riboviria, kingdom Pararnavirae, phylum Artverviricota, class Revtraviricetes, order Ortervirales, family Pseudoviridae; there are three genera (Pseudovirus, Hemivirus, and Sirevirus) and 33 species |
Virion
Morphology
As part of their replication cycle, pseudovirids form intracellular virus-like particles (VLPs). These particles do not display infectivity according to the traditional virological definition and remain intracellular (Tucker and Garfinkel 2016). However, there is significant evidence that the VLPs are essential intermediates in the life cycle of pseudovirids (Boeke and Sandmeyer 1991, Garfinkel et al., 1985, Mellor et al., 1985). Members of the family Pseudoviridae are typified by somewhat irregularly shaped VLPs of different diameters (around 30−40 nm) that are round to ovoid, often with electron-dense centers (Burns et al., 1992) Although these VLPs are irregular in their native state, expression of truncated forms of the major structural protein (Gag), yields isometric icosahedral VLPs with a mean radius of 20 nm built on the T=3 or T=4 lattice (Palmer et al., 1997) (Figure 1. Pseudoviridae). Saccharomyces cerevisiae Ty1 virus (SceTy1V) and Drosophila melanogaster copia virus (DmecopiaV) both produce similar-looking particles, but SceTy1V virions are cytoplasmic, whereas those of DmecopiaV are found in the nucleus (Bachmann et al., 2004).
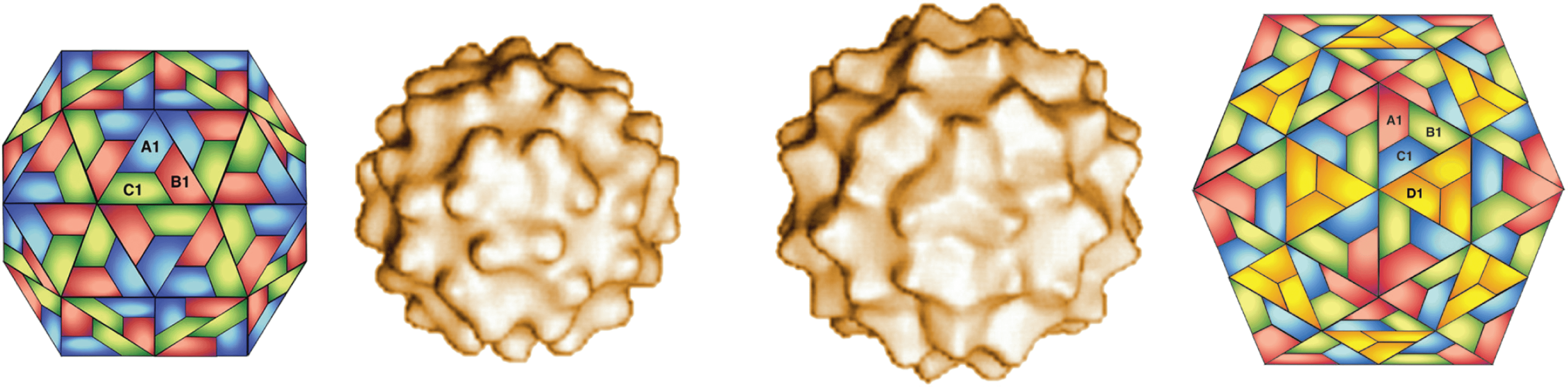 |
| Figure 1. Pseudoviridae. Saccharomyces cerevisiae Ty1 virus particles formed from truncated capsid protein (aa 1−381); surface structure of two forms (T=3, left; T=4, right) of around 30–40 nm determined by cryo-electron microscopy, flanked by the corresponding schematic models (Courtesy of H. Saibil, adapted from (Palmer et al., 1997) with permission from American Society for Microbiology). |
Nucleic acid
Based on current evidence and by analogy to retroviruses, VLPs encapsidate two identical copies of the RNA genome which, depending on the Pseudoviridae species, is 4–10 kb (Figure 2. Pseudoviridae). The genomic RNA is of positive polarity, capped at the 5´-end and polyadenylated at the 3´-end. In addition to the RNA genome, some cellular RNAs, such as specific tRNAs involved in reverse transcription, are also packaged into the VLPs. The genome of members of the family Pseudoviridae presents in two nucleic acid states: the DNA provirion genome and the RNA genome. In its provirion state, a canonical pseudovirid element consists of a DNA sequence of variable size (4 to 10 kbp) inserted in the genome of its host. The RNA genome consists of a full-length (LTR-to-LTR) transcript.
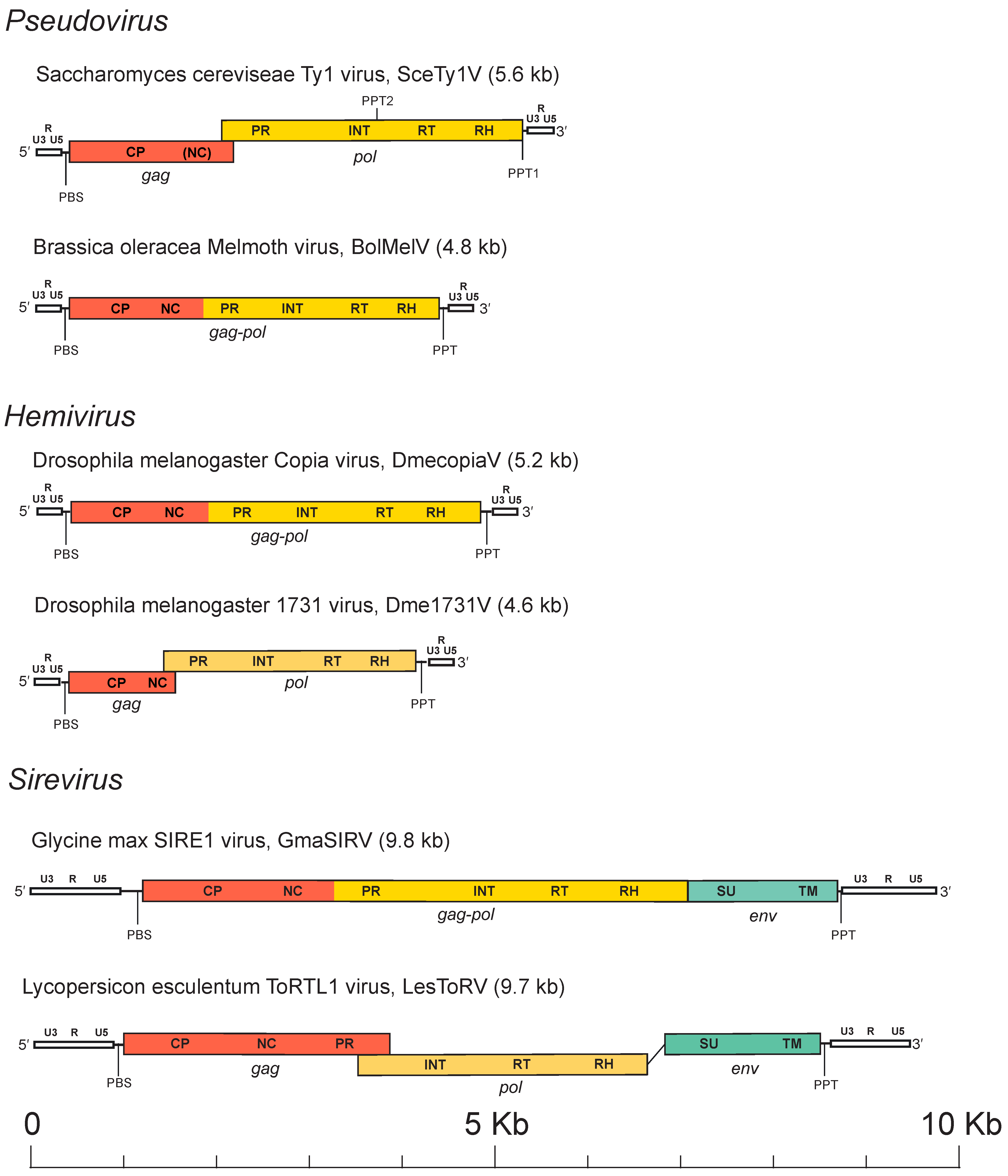 |
| Figure 2. Pseudoviridae. Pseudovirid genome architectures of representative members of the three genera. LTRs are white and show labels for the U3, R and U5 regions. |
Proteins
The icosahedral virions are formed from the Gag polyprotein, which contains the capsid and nucleocapsid domains homologous to the corresponding proteins of retroviruses and other members of the order Ortervirales (Krupovic and Koonin 2017). The capsid (CA) protein is involved in forming the icosahedral shell, whereas the nucleocapsid (NC) protein is an RNA-binding protein which plays a role in the packaging of the genomic RNA into the VLP. As observed in other members of the Ortervirales, the NC of some pseudovirids may present one or more zinc finger motifs (Cys-X2-Cys-X4-His-X4-Cys) at the C-terminus, as in the case of the Saccharomyces cerevisiae Ty4 virus (SceTy4V), or none, as in Saccharomyces cerevisiae viruses Ty1 and Ty2 (SceTy1V and SceTy2V, respectively) (Peterson-Burch and Voytas 2002, Llorens et al., 2009).
Lipids
None present.
Carbohydrates
None present.
Genome organization and replication
A canonical member of the Pseudoviridae has a genome with two genes, gag and pol, which are typical of all members of the Ortervirales (Krupovic et al., 2018). The pol gene is usually expressed at lower levels than gag. In some pseudovirids, Gag and Pol proteins are encoded by a single open reading frame (ORF), whereas in others the two ORFs are separated by a frame-shift or by a stop codon. Pol encodes enzymes required for genome replication (Peterson-Burch and Voytas 2002), namely, protease (PR), integrase (INT) and a reverse transcriptase (RT) with the associated ribonuclease H (RH) subdomain. PR, the first domain encoded by pol, is involved in processing of Gag and is also needed to release the other enzymes from the Pol precursor. INT is characterized by three domains: the HHCC domain, the catalytic core (DD35E motif), and a poorly conserved C-terminal domain (Peterson-Burch and Voytas 2002, Haren et al., 1999). Finally, most (but not all) members of the genus Sirevirus contain a third ORF encoding a polyprotein with features resembling the surface (SU) and transmembrane (TM) protein domains observed in retroviral Env polyproteins (Laten et al., 1998, Kapitonov and Jurka 1999, Peterson-Burch et al., 2000). It is thus possible that sireviruses form infectious extracellular virions, which, however, are yet to be detected and characterized.
The coding region in pseudovirid genomes is flanked by two long terminal repeats (LTRs), which are two identical non-coding DNA sequences. The length of the full genome is variable and may range from 4 kb to more than 9 kb (Figure 2. Pseudoviridae). LTRs are also variable in size. A canonical LTR in members of the family Pseudoviridae presents three regions, namely, U3-R-U5, that are analogous to those of retroviruses (Kumar et al., 1997). By analogy with retroviruses and LTR retrotransposons, U3 contains the promoters; R is repeated on each end of the transcript; and U5 constitutes the first portion of the reverse-transcribed genome. The LTRs do not contain protein-coding genes, but rather regulatory elements (enhancers and promoters) that regulate the expression of the genes found in the internal region of the pseudovirids. The internal region is delimited by two small motifs: one downstream of the 5′-LTR called the primer binding site (PBS), which is usually complementary to the initiator tRNAMet, and by another small region, located upstream of the 3′-LTR, called the polypurine tract (PPT).
The internal region may present one (gag-pol), two (gag and pol) or three (gag, pol and env) ORFs. Whenever detected, the putative envelope proteins are encoded downstream of the RH domain. In almost all members of the Pseudoviridae, the domain architecture is thus inferred to be: 5′-LTR-CA-NC-PR-INT-RT-RH-LTR-3′ or 5′-LTR-CA-NC-PR-INT-RT-RH-SU-TM-LTR-3′. Note that pseudovirids differ from viruses in the other families of the order Ortervirales in the position of their INT domain, which is located between the PR and the RT domains while in members of all other Ortervirales families, it is usually found after the RH domain. The mechanism(s) that regulates Gag and Gag-Pol expression for most single-ORF viruses is unknown. RT-RH mediates the conversion of the full-length genome transcript into a full-length nucleic acid duplex containing the full-length LTR sequences in the form of dsDNA. This DNA is then integrated into the host DNA by INT where it becomes a part of the host genome and can persist there. The integrated form (equivalent to the retroviral provirus) is then transcribed by the host RNA polymerase II to generate new virus RNAs, which are subsequently capped and polyadenylated by host enzymes. The processed transcript is exported to the cytoplasm, where it can be translated into two types of proteins, Gag and Gag-Pol. These proteins co-assemble into an immature virion, which contains RNA and unprocessed polyproteins. Pre-processing of these proteins leads to a change in the virion structure. Proteins encoded in the pol gene (PR, INT, RT-RH) are released from the Gag-Pol precursor and are thought to be activated by pre-processing. Once RT is pre-processed, it converts the RNA molecules to a full-length cDNA which is transported back to the nucleus of the host cell and is inserted into a new site in the host genome, where it becomes permanently fixed (Boeke 2013). In most viruses, the reverse transcription and integration processes closely mimic the replication of retroviruses. The RT and its associated RH subdomain generate a cDNA copy of the LTR retroelement from genomic mRNA (Telesnitsky and Goff 1997).
Biology
The diversity of pseudoviruses and other reverse-transcribing viruses and LTR retrotransposons is now known to be greater than previously thought. Pseudoviruses constitute an intrinsic and significant part of the genome of many eukaryotic species, especially plants. For most of these viruses, the virion is an essential part of the multiplication cycle but is not infectious under normal conditions (in the traditional virological sense). Interestingly, it is very common to find multiple distinct members of one or more pseudovirid species in the genome of the same host organism (for example, SceTy1V, SceTy2V and SceTy4V of S. cerevisiae). However, some of these viruses inhabit the genome of two or more host species, probably because they were already present in their common ancestor. When reverse-transcribing viruses and LTR retrotransposons colonize the germinal lines of their hosts, they are transmitted vertically over generations. In fact, pseudoviruses and all other integrative reverse-transcribing viruses (particularly metaviruses) are excellent molecular markers of evolution in eukaryotes (Llorens et al., 2009). During retrotransposition, the double-stranded proviral cDNA that has been synthetized in the VLP is imported into the nucleus and then inserted into a chromosomal target site. The location and distribution of pseudoviruses in their host genomes varies. Depending on the insertion site, the integration can be mutagenic if it disrupts or alters gene functions, with potential detrimental effects on the viability of host cells and, by extension, viability of the inserted virus. In the course of evolution, pseudovirids (and other retroelements) have developed mechanisms to specifically target the integration site without altering the gene functions. This is primarily achieved by integration into noncoding regions, preferential targeting of heterochromatin regions (not permissive for transcription) or by association with centromeric regions. For example, SceTy1V, SceTy2V and SceTy4V are preferentially inserted into euchromatic regions of S. cerevisiae, near genes transcribed by RNA polymerase III, while Saccharomyces cerevisiae Ty5 virus (SceTy5V) inserts preferentially into subtelomeric heterochromatin. In the case of Drosophila pseudovirids, integration preferentially takes place into euchromatic regions and not necessarily near genes transcribed by RNA polymerase III. In plants, pseudovirids are usually located in the euchromatin, with some exceptions, for example, the onion Allium cepa, wherein they show stronger representation in the heterochromatin than in euchromatin (Flavell et al., 1997).
Derivation of names
Hemivirus: from Greek hemi for “half”, referring to the half-molecule of tRNA used as a primer for reverse transcription.
Pseudovirus: from Greek pseudo for “false”, to connote an evolutionary relationship to viruses with extracellular virions.
Sirevirus: from the abbreviation of the species name: Glycine max SIRE1 virus
Genus demarcation criteria
The family Pseudoviridae belongs to the order Ortervirales (Krupovic et al., 2018). The three genera – Pseudovirus, Hemivirus and Sirevirus – were originally established based on the different length of the tail of the tRNA molecule that is used as a primer to initiate the reverse transcription. Hemiviruses use only a short segment of tRNA in comparison to members of the genus Pseudovirus (Bousios and Darzentas 2013). In addition, sireviruses are found exclusively in plants, and were classified as a separate genus because the first characterized sireviruses encode an additional ORF downstream of pol, reminiscent of the retroviral env genes. This criterion for genus demarcation is under revision as it is inconsistent with the evolutionary history of the Pseudoviridae family. Members of both the Pseudovirus and Hemivirus genera form polyphyletic branches in all Ty1/Copia phylogenies and the current taxonomic structure is insufficient to encompass the diversity observed within the family. Further updates in the genus demarcation of pseudovirids are expected to be based on phylogenetic criteria.
Species demarcation criteria in the family
At present, viruses in the family Pseudoviridae are considered to belong to separate species if at least one of the major coding regions (e.g. capsid) is <50% identical to each other. For example, Ty1 and Ty2 Gag aa sequences are 49% identical and belong to different species in the genus Pseudovirus. The RT domain has also been used for classification (Xiong and Eickbush 1990), with members of different species being <90−95% identical, although the ranges between inter-species and intra-species comparisons may overlap. The host species cannot be used for virus species demarcation as members of multiple species can be present in the same host; for example, SceTy1V, SceTy2V, SceTy4V and SceTy5V are all present in the genome of S. cerevisiae (Havecker et al., 2004, Kumar and Bennetzen 1999, Voytas and Boeke 2002), but belong to four different species in two different genera.
Relationships within the family
The amino acid sequences of the RT, RH and INT proteins are widely used to infer the phylogeny of reverse-transcribing viruses due to their strong and consistent phylogenetic signal. Similar phylogenies are also obtained for the Gag and PR proteins even though they show high divergence among members of the same family (Llorens et al., 2009, Llorens et al., 2008, Llorens et al., 2011). Based on analysis of the most conserved part of the RT domain, two of the three genera are polyphyletic (Figure 3. Pseudoviridae). The Pseudoviridae family splits into at least 16 phylogenetic clades that are named based on one representative from each clade. Two clades, designated as Copia and 1731 are specifically found in drosophilids, while four clades are present in the genomes of plants, designated as Tork, Retrofit, Oryco and Sire. The later, collects all sireviruses in a single clade corresponding to the genus Sirevirus and that supports the perspective of a monophyletic relationship to exist among all members of this genus. Fungi also have diverse pseudovirid populations, with three clades, designated pCretro, Ty1/Tse, and Ty5/Tca (Figure 3. Pseudoviridae). As also observed with the Metaviridae family, members of the family Pseudoviridae are widely distributed in marine animals; this is in contrast to land animals, where members of the Retroviridae predominate. The clade designated as Hydra seems to be specific to fishes; Osser clade groups distinct pseudovirids of algae; GalEA includes pseudovirids from urochordates, fishes and crustaceans; finally, CoDi-C, CoDi-D, CoDi-I and CoDi-II are four lineages from diatoms (Maumus et al., 2009). Several other pseudovirids constitute single-sequence clades in the phylogeny. These are the Zea mays Hopscotch virus (ZmaHopV), Physarum polycephalum Tp1 virus (PpoTp1V), Aedes aegypti Mosqcopia virus (AaeMostV), as well as several unclassified elements such as Porphyra yezoensis PyRE1G1 virus, Bombyx mori Yokozuna virus, Heliconius numata Humnum virus, Tribolium castaneum Tricopia virus, the Anopheles gambiae Mtanga virus, and Oryza sativa Oryco1-2 virus maHopV. For more details, the Gypsy Database (GyDB http://gydb.org) provides online access to a collection of phylogenetic trees for all reverse-transcribing viruses inferred based on the distinct Gag and Pol (and Env) protein domains.
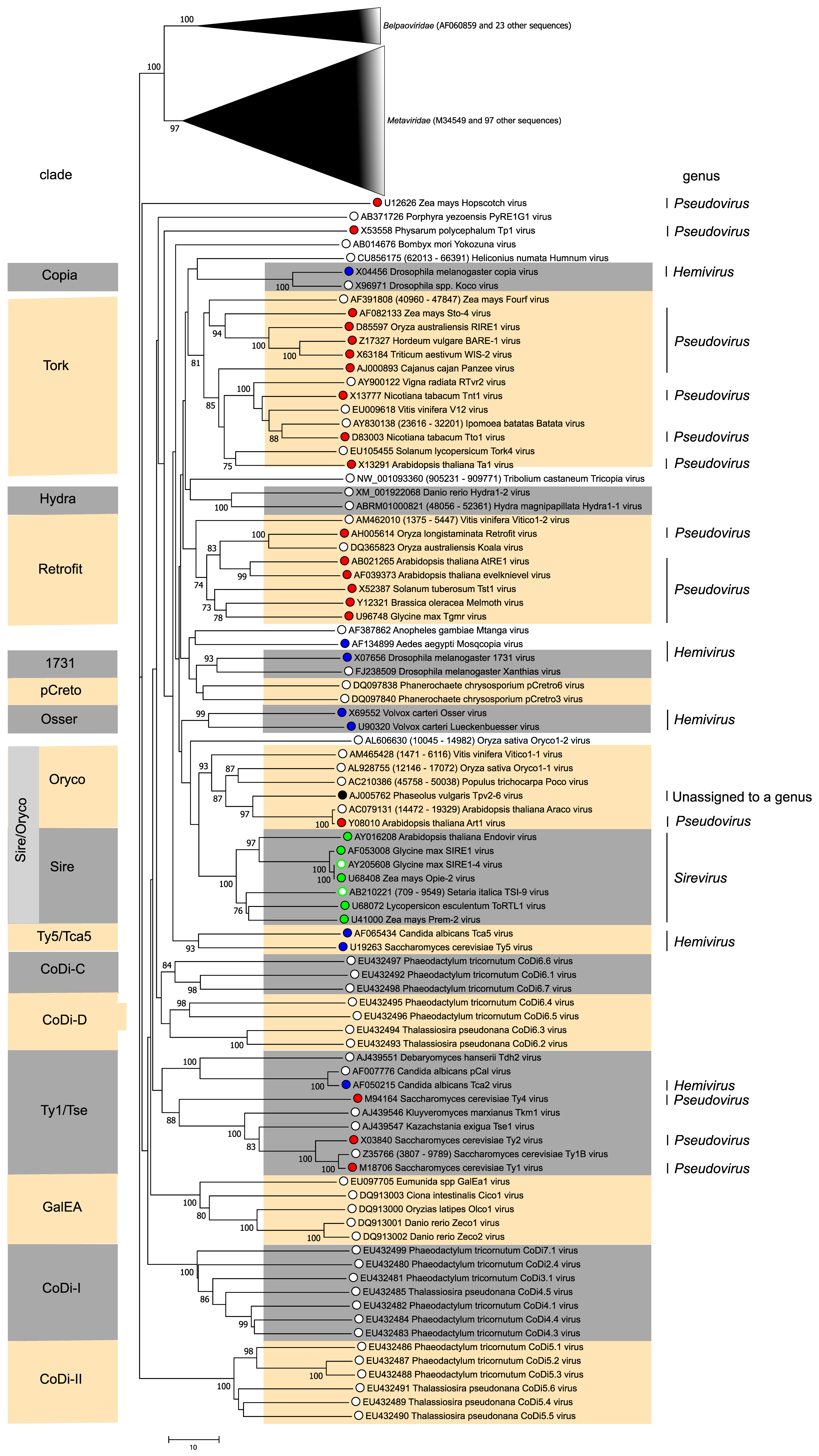 |
| Figure 3. Pseudoviridae. Phylogenetic tree of members of the Pseudoviridae family based on the alignment of the RT core obtained from 210 classified viruses belonging to the Pseudoviridae, Metaviridae and Belpaoviridae families and related unclassified viruses. The alignment was created using Clustal W (Larkin et al., 2007) and GeneDoc (https://genedoc.software.informer.com/2.7) to respectively align and manually refine the sequences. Clustal W and the Neighbor Joining method of phylogenetic reconstruction and 1000 bootstrap replicates were used to infer the tree. Branches for viruses in the Metaviridae and Belpaoviridae families are collapsed, these families being used to root the tree. Bootstrap support is provided where >50% (1000 replicates). Coloured dots at tips indicate viruses in species assigned to the genera Hemivirus (blue), Pseudovirus (red) and Sirevirus (green); a black dot indicates a virus in a species unassigned to a genus, and open dots indicate related, unclassified viruses. Clades based on the analysis of both Gag and Pol proteins are indicated to the left and by shading and follow information provided at GyDB. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Members of the Pseudoviridae family have a shared evolutionary history with members of the other families in the order Ortervirales, including Metaviridae, Belpaoviridae, Retroviridae and Caulimoviridae (Llorens et al., 2020). The minimal conserved core genome of viruses in the families Metaviridae, Pseudoviridae and Belpaoviridae displays the “LTR-gag-pol-LTR” genomic architecture, although pseudovirids differ from the other families in that they have an unusual PR-IN-RT-RH organization of the pol gene. However, some members of the three families have been shown to carry an additional env gene, located downstream of pol (“LTR-gag-pol-env-LTR”), a genomic architecture typical of simple retroviruses that lack accessory genes. The four families of reverse transcribing viruses containing LTRs are also related to viruses of the family Caulimoviridae, which have dsDNA genomes and infect plants. The genomes of caulimovirids usually contain two ORFs encoding coat (gag) and pol polyproteins, with domain features closely similar to those of the other members of the Ortervirales. This structural similarity is also supported by phylogenetic analyses based on sequences of the shared proteins/domains.
Species unsassigned to a genus
The species Phaseolus vulgaris Tpv2-6 virus is currently unassigned to a genus. Close homologs of Phaseolus vulgaris Tpv2-6 virus (PvuTpvV) have been identified in the genomes of a variety of plant species. Current phylogenies show that PvuTpvV and their relatives constitute a well-supported clade close to sireviruses and designated as Oryco clade (Figure 3. Pseudoviridae).
The Member Species table enumerating important virus exemplars classified under each species of the genus is provided at the bottom of the page.
Related, unclassified viruses
| Clade$ | Virus name | Accession number | Coordinates* |
| 1731 | Drosophila melanogaster Xanthias virus | FJ238509 | |
| CoDi-C | Phaeodactylum tricornutum CoDi6.1 virus | EU432492 | |
| CoDi-C | Phaeodactylum tricornutum CoDi6.6 virus | EU432497 | |
| CoDi-C | Phaeodactylum tricornutum CoDi6.7 virus | EU432498 | |
| CoDi-D | Phaeodactylum tricornutum CoDi6.4 virus | EU432495 | |
| CoDi-D | Phaeodactylum tricornutum CoDi6.5 virus | EU432496 | |
| CoDi-D | Thalassiosira pseudonana CoDi6.2 virus | EU432493 | |
| CoDi-D | Thalassiosira pseudonana CoDi6.3 virus | EU432494 | |
| CoDi-I | Phaeodactylum tricornutum CoDi2.4 virus | EU432480 | |
| CoDi-I | Phaeodactylum tricornutum CoDi3.1 virus | EU432481 | |
| CoDi-I | Phaeodactylum tricornutum CoDi4.1 virus | EU432482 | |
| CoDi-I | Phaeodactylum tricornutum CoDi4.3 virus | EU432483 | |
| CoDi-I | Phaeodactylum tricornutum CoDi4.4 virus | EU432484 | |
| CoDi-I | Phaeodactylum tricornutum CoDi4.5 virus | EU432485 | |
| CoDi-I | Phaeodactylum tricornutum CoDi7.1 virus | EU432499 | |
| CoDi-II | Phaeodactylum tricornutum CoDi5.1 virus | EU432486 | |
| CoDi-II | Phaeodactylum tricornutum CoDi5.2 virus | EU432487 | |
| CoDi-II | Phaeodactylum tricornutum CoDi5.3 virus | EU432488 | |
| CoDi-II | Thalassiosira pseudonana CoDi5.4 virus | EU432489 | |
| CoDi-II | Thalassiosira pseudonana CoDi5.5 virus | EU432490 | |
| CoDi-II | Thalassiosira pseudonana CoDi5.6 virus | EU432491 | |
| Copia | Drosophila spp. Koco virus | X96971 | |
| GalEA | Ciona intestinalis Cico1 virus | DQ913003# | |
| GalEA | Danio rerio Zeco1 virus | DQ913001# | |
| GalEA | Danio rerio Zeco2 virus | DQ913002# | |
| GalEA | Eumunida spp GalEa1 virus | EU097705 | |
| GalEA | Oryzias latipes Olco1 virus | DQ913000# | |
| Hydra | Danio rerio Hydra1-2 virus | XM_001922068 | |
| Hydra | Hydra magnipapillata Hydra1-1 virus | ABRM01000821 | (48056 – 52361) |
| Oryco | Arabidopsis thaliana Araco virus | AC079131 | (14472 – 19329) |
| Oryco | Oryza sativa Oryco1-1 virus | AL928755 | (12146 – 17072) |
| Oryco | Populus trichocarpa Poco virus | AC210386 | (45758 – 50038) |
| Oryco | Vitis vinifera Vitico1-1 virus | AM465428 | (1471 – 6116) |
| pCreto | Phanerochaete chrysosporium pCretro3 virus | DQ097840 | |
| pCreto | Phanerochaete chrysosporium pCretro6 virus | DQ097838 | |
| Retrofit | Oryza australiensis Koala virus | DQ365823 | |
| Retrofit | Vitis vinifera Vitico1-2 virus | AM462010 | (1375 – 5447) |
| Tork | Ipomoea batatas Batata virus | AY830138 | (23616 – 32201) |
| Tork | Solanum lycopersicum Tork4 virus | EU105455 | |
| Tork | Vigna radiate RTvr2 virus | AY900122 | |
| Tork | Vitis vinifera V12 virus | EU009618 | |
| Tork | Zea mays Fourf virus | AF391808 | (40960 – 47847) |
| Ty1/Tse | Candida albicans pCal virus | AF007776 | |
| Ty1/Tse | Debaryomyces hanserii Tdh2 virus | AJ439551 | |
| Ty1/Tse | Kazachstania exigua Tse1 virus | AJ439547 | |
| Ty1/Tse | Kluyveromyces marxianus Tkm1 virus | AJ439546 | |
| Ty1/Tse | Saccharomyces cerevisiae Ty1B virus | Z35766 | (3807 – 9789) |
| Anopheles gambiae Mtanga virus | AF387862 | ||
| Bombyx mori Yokozuna virus | AB014676 | ||
| Heliconius numata Humnum virus | CU856175 | (62013 – 66391) | |
| Oryza sativa Oryco1-2 virus | AL606630 | (10045 – 14982) | |
| Porphyra yezonsis PyRE1G1 virus | AB371726 | ||
| Tribolium castaneum Tricopia virus | NW_001093360 | (905231 – 909771) |
Virus names and virus abbreviations are not official ICTV designations.
$ Clades assignment according to GyDB.
# Partial genome sequence
* Numbers in parentheses are the coordinates of the virus within a larger host sequence

