Family: Bicaudaviridae
David Prangishvili and Mart Krupovic
The citation for this ICTV Report chapter is the summary published as Prangishvili et al., (2018):
ICTV Virus Taxonomy Profile: Bicaudaviridae, Journal of General Virology, 99: 864–865.
Corresponding authors: David Prangishvili ([email protected]) and Mart Krupovic ([email protected])
Edited by: Andrew M. Kropinski and Stuart G. Siddell
Posted: May 2018
PDF: ICTV_Bicaudaviridae.pdf
Summary
The family Bicaudaviridae includes viruses that infect hyperthermophilic archaea in the genus Acidianus (Table 1 Bicaudaviridae). The circular double-stranded DNA genome of Acidianus two-tailed virus consists of 62,730 bp, and replication can be either lytic or lysogenic. The virions undergo unique extracellular morphogenesis. Virions are released from host cells as spindle-shaped particles, which subsequently develop long tails, one at each of the two pointed ends. The spindle-shaped morphology is unprecedented among viruses of bacteria and eukaryotes and represents a group of archaea-specific virion morphotypes.
Table 1 Bicaudaviridae. Characteristics of members of the family Bicaudaviridae.
| Characteristic | Description |
| Typical member | Acidianus two-tailed virus (AJ888457), species Bicaudavirus pozzuoliense |
| Virion | Spindle-shaped upon release (120 x 80 nm), subsequently developing two tails each of up to 400 nm in length |
| Genome | Circular dsDNA of 62,730 bp |
| Replication | Lytic or lysogenic |
| Translation | Not known |
| Host range | Hyperthermophilic archaea from the genus Acidianus |
| Taxonomy | One genus including a single species |
Virion
Morphology
Virions of Acidianus two-tailed virus (ATV) are released from host cells as spindle-shaped particles, about 120×80 nm, which subsequently develop two long tails, one at each of the two pointed ends (Table 1 Bicaudaviridae, Figure 1 Bicaudaviridae) (Häring et al., 2005, Prangishvili et al., 2006). The tails are heterogeneous in length, reaching 400 nm. The tails have a tube-like structure with walls that are approximately 6 nm thick. The tube terminates with a narrow channel, which is 2 nm in width, and a terminal anchor-like structure formed by two furled filaments, each with a width of 4 nm.
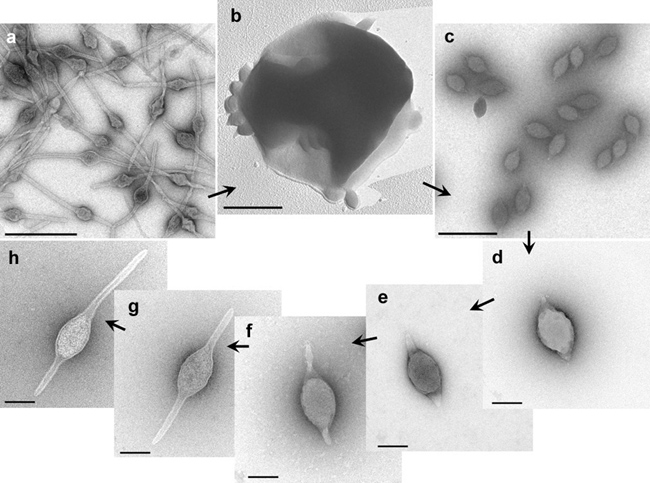 |
| Figure 1 Bicaudaviridae. Electron micrographs of different forms of Acidianus two-tailed virus. a, Virions in the infected cell culture at the late stage of tail development. b, extrusion of virions from an Acidianus convivator cell; c, virions in growing culture of infected A. convivator, two days post-infection; d, as for c, but purified by CsCl gradient centrifugation; e-h, as for d, but after incubation at 75°C for 2, 5, 6, and 7 days, respectively. Samples were negatively-stained with 3% uranyl acetate, except for b, which was platinum- shadowed. Bars: a-c, 0.5 µm; d-h, 0.1 µm (first published in (Häring et al., 2005)). |
Physicochemical and physical properties
Extracellular morphological development of the virion takes place specifically at temperatures above 75 °C, close to that of the natural habitat, and does not require the presence of the host cells, an exogenous energy source or specific co-factors. The average volume of the tailless virion is about 1.4 nm3, and that of the two-tailed virion 6.2 nm3. However, the total surface areas for the two types of virions are similar, about 6×103 nm2. The mechanism of ATV tail development is unknown. The buoyant density of ATV virions in CsCl is about 1.3 g cm-3.
Nucleic acid
Virions of ATV contain one molecule of circular dsDNA of 62,730 bp, with a GC content of 41.2%.
Proteins
Protein patterns of tailless and two-tailed virions are identical. The virions carry 11 major structural proteins (90, 80, 70, 60, 48, 45, 34, 22, 16, 14 and 12 kDa; Table 2.Bicaudaviridae), five of which are modified at their N-termini. Several of the larger proteins are rich in coiled-coil or low complexity sequence domains, or both. The 80 kDa protein appears to be modified in the two-tailed but not in the tail-less virions (Prangishvili et al., 2006). High resolution structures of 32 and 14 kDa proteins have been solved by X-ray crystallography (Felisberto-Rodrigues et al., 2012, Goulet et al., 2010) and both display unique folds that are not observed in proteins of other classified viruses (Krupovic et al., 2018). The structural protein encoded by ORF140 (Table 2 Bicaudaviridae) is a paralog of the major capsid protein encoded by ORF131. It has been shown that the product of ORF140 acts as a global transcription repressor, which forms a high-affinity complex with the host RNA polymerase by binding inside the DNA-binding channel. This interaction counteracts the formation of transcription pre-initiation complexes in vitro and represses transcription initiation as well as elongation (Sheppard et al., 2016). However, the role of ORF140 in transcription regulation in vivo, as well as in virion assembly and structure, remain to be investigated. ATV structural proteins encoded by ORF387, ORF653 and ORF800 as well as the non-structural protein containing the von Willebrand factor type A domain (encoded by ORF618) have been shown to interact with the MoxR-like AAA+ ATPase, which is also a structural virion component (Scheele et al., 2011). It has been suggested that the latter group of proteins may constitute a chaperone system for extracellular tail development.
Lipids
No lipids were obtained from ATV virions after treatment with chloroform/methanol (1:1, vol/vol).
Carbohydrates
Not known. Note that the virus encodes a putative glycosyltransferase.
Genome organization and replication
The ATV genome encodes 72 predicted proteins and carries four putative transposable elements (Figure 2. Bicaudaviridae, Table 2.Bicaudaviridae). Forty-three genes are predicted to produce leader-less transcripts and 35 genes are organized in 12 putative operons. Several genes may have been produced by gene duplication. Besides structural virion proteins, the virus encodes several functionally diverse enzymes, including a putative integrase of the tyrosine recombinase superfamily, a glycosyltransferase, a ParB-like partitioning protein, a DNA repair photolyase, an acyltransferase and three distinct AAA+ ATPases. For two of the ATPases (ORF529 and ORF618), the activity has been demonstrated in vitro (Scheele et al., 2011, Erdmann et al., 2011).
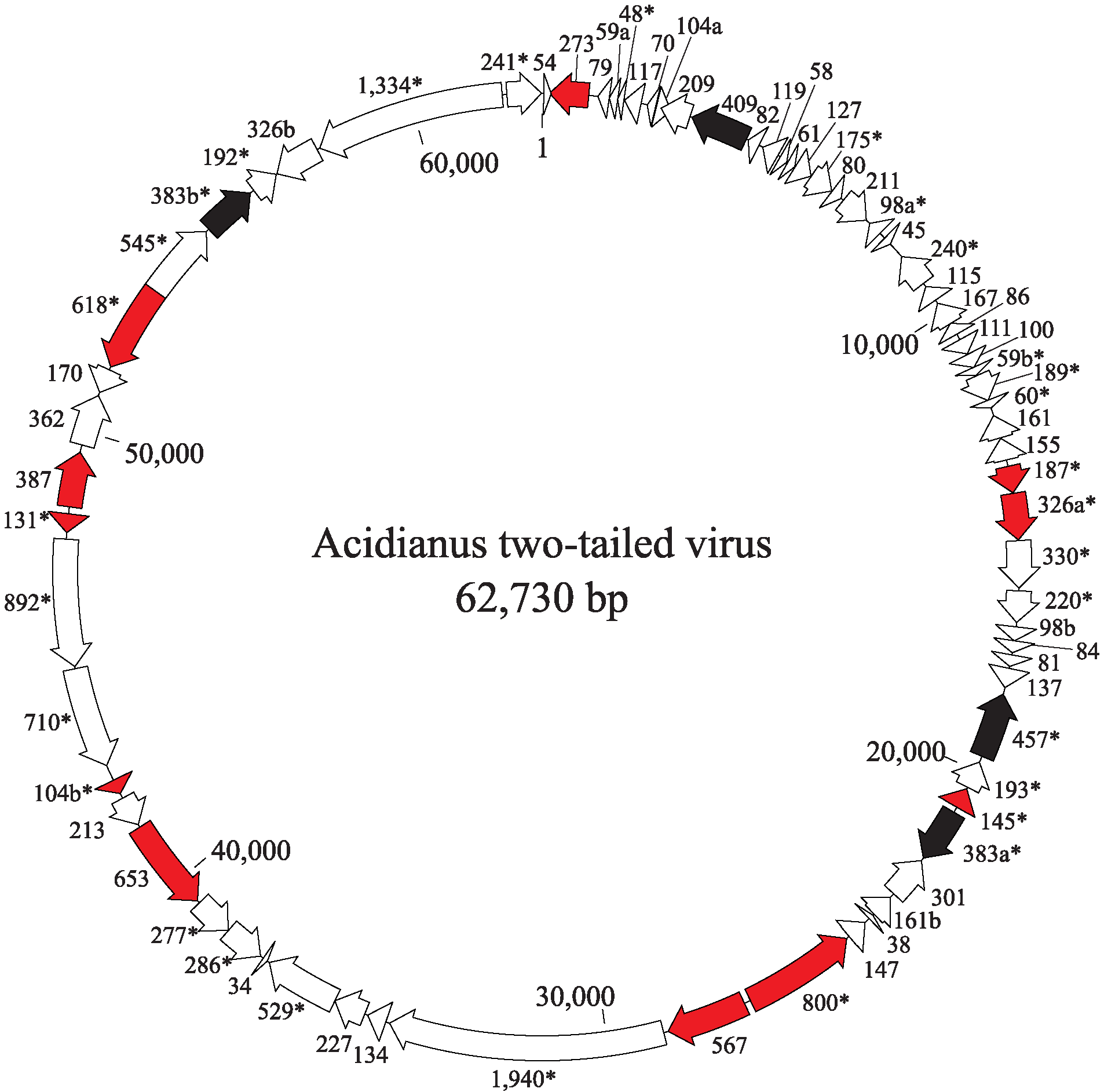 |
| Figure 2 Bicaudaviridae. Genome organization of Acidianus two-tailed virus showing location, sizes and transcriptional direction of the putative genes. Color-coded ORFs correspond to: red, virion proteins; black, transposable elements. ORFs shared with unclassified large spindle-shaped viruses are indicated with asterisks. |
Table 2 Bicaudaviridae. Genome annotation of Acidianus two-tailed virus.
| ORF | Protein Accession Number | Gene product | Function |
| ORF54 | YP_319832 | gp01 | |
| ORF273 | YP_319833 | gp02 | Structural protein (PDB id: 4art) |
| ORF79 | YP_319834 | gp03 | |
| ORF59a | YP_319835 | gp04 | |
| ORF48 | YP_319836 | gp05 | |
| ORF117 | YP_319837 | gp06 | Putative DNA-binding protein |
| ORF70 | YP_319838 | gp07 | Transcription regulator, winged helix-turn-helix (wHTH) domain |
| ORF104a | YP_319839 | gp08 | |
| ORF209 | YP_319840 | gp09 | |
| ORF409 | YP_319841 | gp10 | TnpB protein associated with IS200/IS605 transposons |
| ORF82 | YP_319842 | gp11 | |
| ORF119 | YP_319843 | gp12 | |
| ORF58 | YP_319844 | gp13 | |
| ORF61 | YP_319845 | gp14 | |
| ORF127 | YP_319846 | gp15 | |
| ORF175 | YP_319847 | gp16 | |
| ORF80 | YP_319848 | gp17 | |
| ORF211 | YP_319849 | gp18 | |
| ORF98a | YP_319850 | gp19 | Transcription regulator, wHTH domain |
| ORF45 | YP_319851 | gp20 | |
| ORF240 | YP_319852 | gp21 | |
| ORF115 | YP_319853 | gp22 | |
| ORF167 | YP_319854 | gp23 | |
| ORF86 | YP_319855 | gp24 | RNA-binding Lsm/Hfq-like protein |
| ORF111 | YP_319856 | gp25 | |
| ORF100 | YP_319857 | gp26 | |
| ORF59b | YP_319858 | gp27 | Transcription regulator, RHH domain |
| ORF189 | YP_319859 | gp28 | Transcription regulator, C2H2-type zinc finger |
| ORF60 | YP_319860 | gp29 | Transcription regulator, RHH domain |
| ORF161 | YP_319861 | gp30 | |
| ORF155 | YP_319862 | gp31 | |
| ORF187 | YP_319863 | gp32 | Structural protein |
| ORF326a | YP_319864 | gp33 | Structural protein |
| ORF330 | YP_319865 | gp34 | Acyltransferase, peptidoglycan/LPS O-acetylase |
| ORF220 | YP_319866 | gp35 | |
| ORF98b | YP_319867 | gp36 | Transcription regulator, C2H2-type zinc finger |
| ORF84 | YP_319868 | gp37 | |
| ORF81 | YP_319869 | gp38 | |
| ORF137 | YP_319870 | gp39 | |
| ORF457 | YP_319871 | gp40 | TnpB protein associated with IS200/IS605 transposons |
| ORF193 | YP_319872 | gp41 | |
| ORF145 | YP_319873 | gp42 | Structural protein, paralog of the major structural protein; transcriptional repressor |
| ORF383a | YP_319874 | gp43 | TnpB protein associated with IS200/IS605 transposons |
| ORF301 | YP_319875 | gp44 | DNA repair photolyase, radical SAM superfamily |
| ORF161b | YP_319876 | gp45 | |
| ORF38 | YP_319877 | gp46 | |
| ORF147 | YP_319878 | gp47 | |
| ORF800 | YP_319879 | gp48 | Structural protein; tetratricopeptide repeat protein |
| ORF567 | YP_319880 | gp49 | Structural protein |
| ORF1940 | YP_319881 | gp50 | |
| ORF134 | YP_319882 | gp51 | |
| ORF227 | YP_319883 | gp52 | |
| ORF529 | YP_319884 | gp53 | AAA+ ATPase |
| ORF34 | YP_319885 | gp54 | |
| ORF286 | YP_319886 | gp55 | DnaA-like AAA+ ATPase |
| ORF277 | YP_319887 | gp56 | |
| ORF653 | YP_319888 | gp57 | Structural protein |
| ORF213 | YP_319889 | gp58 | |
| ORF104b | YP_319890 | gp59 | Structural protein |
| ORF710 | YP_319891 | gp60 | |
| ORF892 | YP_319892 | gp61 | von Willebrand factor type A (vWA) domain |
| ORF131 | YP_319893 | gp62 | Major structural protein (PDB id: 3faj) |
| ORF387 | YP_319894 | gp63 | Structural protein |
| ORF362 | YP_319895 | gp64 | |
| ORF170 | YP_319896 | gp65 | |
| ORF618 | YP_319897 | gp66 | Structural protein; MoxR-like AAA+ ATPase |
| ORF545 | YP_319898 | gp67 | Glycosyltransferase, mannosyltransferase |
| ORF383b | YP_319899 | gp68 | TnpB protein associated with IS200/IS605 transposons |
| ORF192 | YP_319900 | gp69 | |
| ORF326b | YP_319901 | gp70 | ParB-like partitioning protein |
| ORF1334 | YP_319902 | gp71 | |
| ORF241 | YP_319903 | gp72 | Integrase, tyrosine recombinase superfamily |
Biology
The virus was isolated from a hot acidic spring (87–93 °C, pH 1.5–2.0) in Pozzuoli, Italy. The host range is limited to autochthonous species of hyperthermophilic archaea from the genus Acidianus. Infection leads either to viral replication and subsequent cell lysis or conversion of infected cell into a lysogen. The lysogenic cycle involves integration of the viral genome into the host chromosome, probably facilitated by a virus-encoded integrase. The lysogeny can be interrupted by stress factors, e.g. UV-irradiation or a decrease in temperature. The virus does not encode identifiable DNA and RNA polymerases and is likely to depend on the host machineries for genome replication and transcription.
Derivation of names
Bicauda: from Latin bi, “two”, and cauda, “tail”.
Relationships within the family
Putative relatives, sharing a considerable fraction of genes with the bicaudavirus ATV (Figure 2. Bicaudaviridae) have been described, but remain unclassified. These include Acidianus tailed spindle virus (Hochstein et al., 2016), Sulfolobus tengchongensis spindle-shaped virus 1 (Xiang et al., 2005), Sulfolobus tengchongensis spindle-shaped virus 2 (Erdmann et al., 2014) as well as Sulfolobus monocaudaviruses 1 (Erdmann et al., 2013), 2, 3 and 4 (Gudbergsdóttir et al., 2016). Similar to ATV, Sulfolobus monocaudavirus 1 was reported to develop one or two tails extracellularly (Uldahl et al., 2016). By contrast, the other unclassified viruses contain a single long tail, which appears to develop intracellularly. In the future, these viruses are likely to be classified into new genera within the family Bicaudaviridae (Krupovic et al., 2014).
Relationships with other taxa
Bicaudaviruses encode a conserved DnaA-like AAA+ ATPase, which is shared with members of the archaeal virus families Fuselloviridae (Iranzo et al., 2016) and Guttaviridae (Prangishvili et al., 2018), suggesting that the three groups of viruses might be evolutionarily related.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Sulfolobus monocaudavirus 1 | HG322870 | SMV1 |
| Sulfolobus monocaudavirus 2 | KP238129 | SMV2 |
| Sulfolobus monocaudavirus 3 | KP282677 | SMV3 |
| Sulfolobus monocaudavirus 4 | KP282678 | SMV4 |
| Acidianus tailed spindle virus | KU645528 | ATSV |
| Sulfolobus tengchongensis spindle-shaped virus 1 | AJ783769 | STSV1 |
| Sulfolobus tengchongensis spindle-shaped virus 2 | JQ287645 | STSV2 |
Virus names and virus abbreviations are not official ICTV designations.

