Family: Hepadnaviridae
Genus: Avihepadnavirus
Distinguishing features
Duck hepatitis B virus (DHBV) virions are spherical, 46–48 nm in diameter, with a nucleocapsid that is 35 nm in diameter and exhibits projections. Particles without nucleic acids formed by envelope components are pleomorphic and up to 60 nm in diameter. The single-stranded gap in the virion DNA is usually short, about 12 nt. DHBV lacks an X gene containing a conventional initiation codon, but some other avihepadnaviruses may have an X gene with a canonical start codon. Virus particles have only the largest (LHBs, 36 kDa) and smallest (SHBs, 17 kDa) S proteins. Transmission is predominantly vertical. Heron hepatitis B virus (HHBV) differs from DHBV in that a highly conserved ORF is present upstream of the C ORF, in a position analogous to the X gene of orthohepadnaviruses.
Virion
See discussion under family description.
Genome organization and replication
See discussion under family description.
Biology
DHBV is maintained in domestic duck flocks through vertical transmission from viremic ducks. The virus infects the developing liver in ovo and is not recognized sufficiently by the host immune response to produce hepatitis and liver disease, or to eliminate the virus. Liver cancer has not been associated with chronic infection. Transmission to neonates may also occur, leading to chronic infection. Transmission to adults generally leads to transient infection. The biology of HHBV infections in its natural host has not been studied.
Species demarcation criteria
The species demarcation criteria in the genus are:
- More than 20% divergence at nucleotide level of complete genomes – for example HHBV diverges from DHBV by 21.6% and parrot HBV from DHBV and HHBV by between 22.5 1%.
- Host restriction of infectivity – for example, HHBV cannot be transmitted to ducks.
A number of viruses have been isolated from geese and ducks, with sequences more closely related to that of DHBV than HHBV (Figure 1. Avihepadnavirus). A virus isolated from grey crowned cranes (Balearica regulorum), designated crane hepatitis B virus (CHBV) has been shown to infect primary duck hepatocytes (Prassolov et al., 2003). Viruses classified in the species Duck hepatitis B virus have also been isolated from Sheld goose and Ross goose. A virus closely related to HHBV has been isolated from white storks and designated stork hepatitis B virus (STHBV). Like HHBV, STHBV has a low infectivity for duck hepatocytes. Avihepadnaviruses assigned to another species, Parrot hepatitis B virus, have been isolated from parrots (parrot hepatitis B virus).
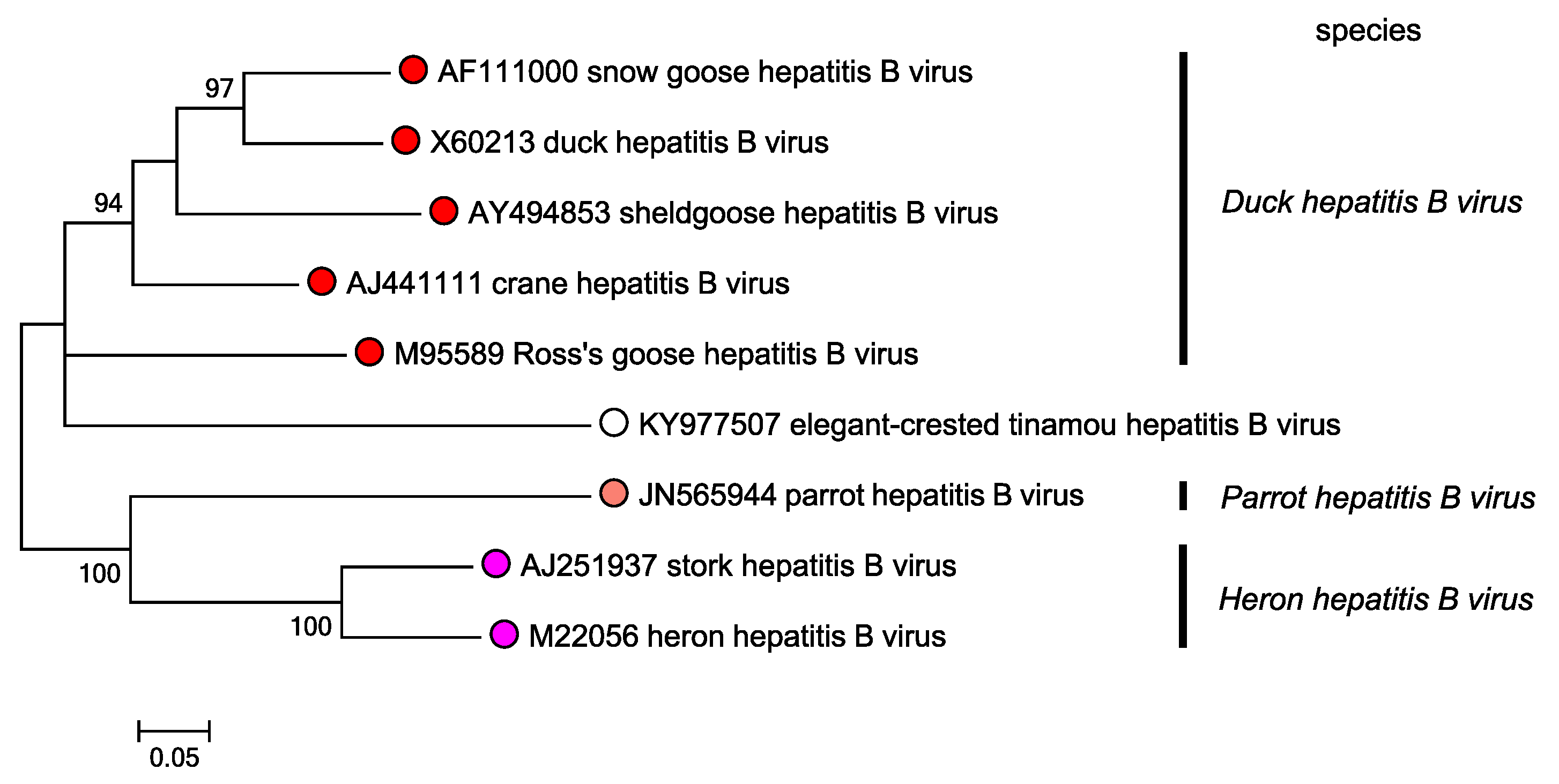 |
|
Figure1. Avihepadnavirus. Phylogenetic tree based on nine avihepadnavirus polymerase gene sequences. Maximum likelihood trees were produced using the General Time Reversible model with a gamma distribution of variation including invariant sites, and conducted using MEGA7 (Kumar et al., 2016). Numbers indicate where bootstrap support for branches was > 70%. Circles at tips are coloured according to species; unclassified viruses have unfilled circles. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Related, unclassified viruses
One highly divergent avihepadnavirus has been isolated from the elegant-crested tinamou, Eudromia elegans, (ETHBV)(Jo et al., 2017).
|
Virus name |
Accession number |
Virus abbreviation |
|
elegant-crested tinamou hepatitis B virus |
ETHBV |
Virus names and virus abbreviations are not official ICTV designations.
Although avihepadnaviruses do not regularly integrate into the host genome, as for herpetohepadnaviruses, fragmented endogenous hepadnaviral elements have been found in the genome of the following birds: zebra finch, designated as zebra finch HBV-derived EVEs (eZHBVs) and other passerines (Gilbert and Feschotte 2010) as well as in the genome of budgerigars (Cui and Holmes 2012). These elements have mutations indicating that they are extinct viruses that have circulated in finch and other avian species in the past. While their presence in avian genomes provides insights into the evolution of viruses in this family, they are not viral genomic sequences and therefore are not to be classified as members of the Hepadnaviridae.

