Family: Ascoviridae
Sassan Asgari, Dennis K. Bideshi, Yves Bigot, Brian A. Federici and Xiao-Wen Cheng
The citation for this ICTV Report chapter is the summary published as Asgari et al., (2017):
ICTV Virus Taxonomy Profile: Ascoviridae, Journal of General Virology, 98:4–5.
Corresponding author: Sassan Asgari (s.asgari@uq.edu.au)
Edited by: Balázs Harrach and Stuart G. Siddell
Posted: December 2016
Summary
The family Ascoviridae includes viruses with circular dsDNA genomes of 100-200 kilobase pairs (kbp) characterized by oblong enveloped virions of 200-400 nm in length (Table 1. Ascoviridae). Members of this family mainly infect lepidopteran larvae and are mechanically transmitted by parasitoid wasps in which they may also replicate. They cause high mortality among economically important insect pests thereby contributing to their natural ability to control insect populations.
Table 1. Ascoviridae. Characteristics of members of the family Ascoviridae.
| Characteristic | Description |
| Typical member | Spodoptera frugiperda ascovirus 1a (AM398843), species Ascovirus sfav1a |
| Virion | Enveloped, 130 nm in diameter by 200-400 nm in length, at least 20 polypeptides |
| Genome | 100-200 kbp of circular dsDNA with 117-180 genes |
| Replication | Nuclear, with cell cleavage into virion-containing vesicles that turn the host haemolymph milky white |
| Translation | From transcribed mRNAs |
| Host Range | Lepidopteran insect larvae, mostly members of the family Noctuidae |
| Taxonomy | Realm Varidnaviria, kingdom Bamfordvirae, phylum Nucleocytoviricota, class Megaviricetes, order Pimascovirales: two genera Ascovirus and Toursvirus with four species |
Members of this family are classified into two genera with distinct evolutionary lineages and genome characteristics:
Ascovirus. This genus includes three species whose members infect various members of the insect family Noctuidae, many species of which are economically important. Ascoviruses are difficult to transmit orally, and experimental studies as well as field observations indicate virions are mechanically transmitted horizontally by endoparasitic wasps of the families Braconidae and Ichneumonidae (Hymenoptera) through their contaminated ovipositor (Bideshi et al., 2010).
Toursvirus. This genus includes only one species, Toursvirus dptv1a (previously named Diadromus pulchellus toursvirus and Diadromus pulchellus ascovirus 4a). Diadromus pulchellus toursvirus (DpTV) is so far only found in species of the lepidopteran family Yponomeutidae, but it replicates also in its ichneumonid vector, Diadromus pulchellus. Replication in the wasp vector is limited in comparison to the titers produced in the lepidopteran host (Bigot et al., 1997a). In the wasp, DpTV is transmitted vertically when the virus genome is carried as unintegrated DNA in the nuclei of infected cells.
Virion
Morphology
Virions of ascoviruses are either bacilliform, ovoidal or allantoid in shape, and depending on the species, have complex symmetry, and are large, measuring approximately 130 nm in diameter by 200-400 nm in length. The virion consists of an inner particle surrounded by an outer envelope. The inner particle typically measures 80 x 300 nm and contains a DNA/protein core bounded by an apparent lipid bilayer, the external surface of which bears a distinctive layer of protein subunits. The virion, therefore, appears to contain two lipid membranes, one associated with the inner particle and the other forming the envelope. In negatively stained preparations, virions have a distinctive reticulate appearance thought to result from superimposition of protein subunits on the surface of the internal particle with those in the external envelope (Figure 1 Ascoviridae). Virions of DpTV of the genus Toursvirus have a similar size: about 220 nm long and 150 nm wide. They are multilayered, with two clear 7-nm-thick outer layers and one 15-nm-thick inner layer surrounding an electron-dense core (155-110 nm). However, the flattened rice-grain shape and fragility of the DpTV particles are unlike that of ascoviruses.
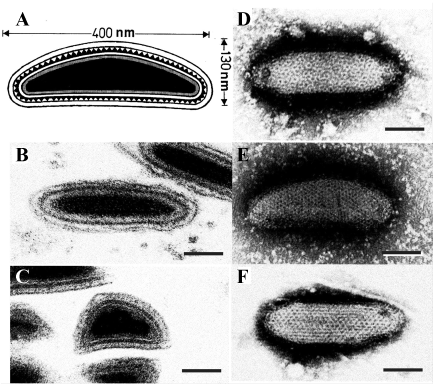 |
| Figure 1 Ascoviridae: Morphology of ascovirus virions. A. Schematic illustration of the structure of a typical ascovirus virion. The virion consists of an inner particle and an outer envelope. The inner particle is complex and contains a DNA/protein core surrounded by an apparent unit membrane, the external surface of which bears a layer of distinctive protein subunits. B, C. Respectively, ultrathin longitudinal- and cross-sections through typical ascovirus virions (SfAV-1a). The dense inner layer corresponds with the distinctive layer of subunits shown in A. D to F. Negatively stained preparations of virions from three different ascovirus species: D, Spodoptera frugiperda ascovirus 1a (SfAV-1a); E, Trichoplusia ni ascovirus 2a (TnAV-2a), and F, Heliothis virescens ascovirus 3a (HvAV-3a). The reticulate appearance of the virions is thought to be due to the superimposition of top and bottom layers of the inner particle and the outer envelope. Bar = 50 nm. |
Physicochemical and physical properties
Virions are sensitive to organic solvents and detergents.
Nucleic acid
The inner particle contains a single molecule of circular dsDNA ranging from 100 to 200 kbp. The G + C content ranges from 42% to 60% depending on the species. Ascovirus genomes contain from 117 to 180 genes, of which 40 are common among them (Bigot et al., 2009).
Proteins
Virions contain at least 20 proteins ranging in size from 6 to 200 kilodaltons (kDa) (Tan et al., 2009).
Lipids
Ultrastructural evidence and detergent sensitivity indicate the presence of lipid in both the outer envelope and inner particle of the virion. The specific lipid composition is not known.
Genome organization and replication
The genomes of five members of the genus Ascovirus, Heliothis virescens ascovirus 3e (HvAV-3e) (Asgari et al., 2007), HvAV-3f (Wei et al., 2014), HvAV-3g (Huang et al., 2012), Spodoptera frugiperda ascovirus 1a (SfAV-1a) (Bideshi et al., 2006) and Trichoplusia ni ascovirus 6a (TnAV-6a; previously named TnAV-2c) (Wang et al., 2006), and one member of the genus Toursvirus, Diadromus wulchellus toursvirus (DpTV) (Bigot et al., 2009) have been completely sequenced.
Members of the family Ascoviridae initiate replication in the nucleus. The nucleus enlarges and ruptures, after which the plasmalemma invaginates, forming internal membranous folds that cleave the cell into a cluster of virion-containing vesicles (Federici and Govindarajan 1990). Virion assembly becomes apparent after the nucleus ruptures. The first recognizable structural component of the virion to form is the multilaminar layer of the inner particle. Based on its ultrastructure, this layer consists of a unit membrane and an exterior layer of protein subunits. As the multilaminar layer forms, the dense DNA/protein core assembles along the inner surface. While this process continues, the allantoid, ovoidal or bacilliform shape of the inner particle becomes apparent. After the inner particle is assembled, it is enveloped by a membrane, synthesized de novo, or derived from cell membranes, within the cell or vesicle. In the case of SfAV-1a, virions are occluded in an occlusion body composed of minivesicles and protein (Federici et al., 1990).
Biology
Host range
Members of the Ascovirus genus cause disease in lepidopteran larvae and pupae, and have been reported most commonly from species of the family Noctuidae, including Trichoplusia ni, Heliothis virescens, Helicoverpa zea, Spodoptera frugiperda and Autographa precationis. TnAV-2a and HvAV-3a have been shown to have a broad experimental host range among larvae of the lepidopteran family Noctuidae, but the host range of SfAV-1a is restricted primarily to species of Spodoptera (Hamm et al., 1986). However, ascoviruses may have an expanded host range that includes non-noctuid insects. For example, recent studies have demonstrated that HvAV-3e is able to productively propagate in Crocidolomia pavonana and Plutella xylostella larva, lepidopteran species belonging to families Crambidae and Plutellidae, respectively (Smede et al., 2008, Furlong and Asgari 2010).
DpTV, the only member of the Toursvirus genus, so far has only been found in species of the lepidopteran family Yponomeutidae, in which it replicates extensively. There is also a limited replication of the virus in its ichneumonid vector, Diadromus pulchellus, but is substantially less than its replication in the lepidopteran host (Bigot et al., 1997a).
Transmission
Members of the family Ascoviridae are difficult to transmit orally, and experimental studies as well as field observations indicate most are transmitted horizontally by endoparasitic wasps (Hymenoptera), many species of which belong to the families Braconidae and Ichneumonidae (Federici and Govindarajan 1990, Tillman et al., 2004). During egg-laying, the ovipositor of female wasps becomes contaminated with virions and virion-containing vesicles that circulate in the blood (hemolymph) of infected caterpillars. Wasps contaminated in this manner subsequently transmit ascovirus virions to new caterpillar hosts during oviposition. DpTV is transmitted vertically in the wasp D. pulchellus when the virus genome is carried as extrachromosomal DNA in the nuclei of infected cells.
Geographical distribution
Members of the family Ascoviridae are known from the United States, Europe, Australia, Indonesia, Japan, China and Mexico, and likely occur worldwide, that is, wherever species of Lepidoptera and their hymenopteran parasites occur. These viruses cause a chronic, fatal disease that markedly retards larval development, but which typically exhibits little other gross pathology. This lack of easily recognizable gross pathology probably accounts for the lack of host records from many geographical regions.
Cytopathic effects
Members of the family Ascoviridae vary in tissue tropism, with some attacking most host tissues, such as TnAV-2a and HvAV-3a, whereas others, such as SfAV-1a, are restricted to the fat body (Federici and Govindarajan 1990). The unique property of ascovirus infection is a cytopathology in which host cells cleave to form virion-containing vesicles by a developmental process utilizing a modified form of apoptosis (Bideshi et al., 2005). Infection results in nuclear hypertrophy followed by lysis. The anucleate cell enlarges 5–10-fold and then cleaves into 10–30 virion-containing vesicles. Membranes delimiting vesicles form by invagination of the plasmalemma and membrane synthesis. Millions of vesicles (ca. 107–108 vesicles ml-1) accumulate in the hemolymph, turning it milky white. Opaque white hemolymph containing refractile virion vesicles is unique and diagnostic for ascovirus disease (Federici 1983).
Derivation of names
Asco: from the Greek for “sac”, referring to the virion-containing vesicles produced by cleavage of host cells, which are characteristic for all known viruses of this family.
Tours: from Tours, France, where DpTV was first isolated.
Genus demarcation criteria
The following list of criteria is used to differentiate genera in the family:
- Virion morphology
- Phylogenetic analysis
- Lack of DNA/DNA hybridization at low stringency
- Host of isolation and experimental host range
- Association with specific hymenopteran parasites, if apparent
The virions of most Ascovirus isolates are similar in size and shape (Federici et al., 1990), but the Toursvirus DpTV virions are more flattened rice-grain shape rather than allantoid found in other ascoviruses (Bigot et al., 1997b). There should be no DNA/DNA hybridization between members of the genera Ascovirus and Toursvirus. DpTV, as the sole member of Toursvirus, is mostly transmitted by the parasitoid wasp, Diadromous pulchellus, whereas members of the genus Ascovirus can be transmitted by a variety of parasitoid wasp species. DpTV has been mainly isolated from the host pupae in the lepidopteran family of Yponomeutidae, but members of the Ascovirus genus have been isolated mostly from larvae of the lepidopteran family Noctuidae.
Relationships within the family
The family appears to have evolved into two lineages in which DpTV sits within a lineage distinct to that of the Ascovirus genus (Piegu et al., 2015) (Figure 2 Ascoviridae).
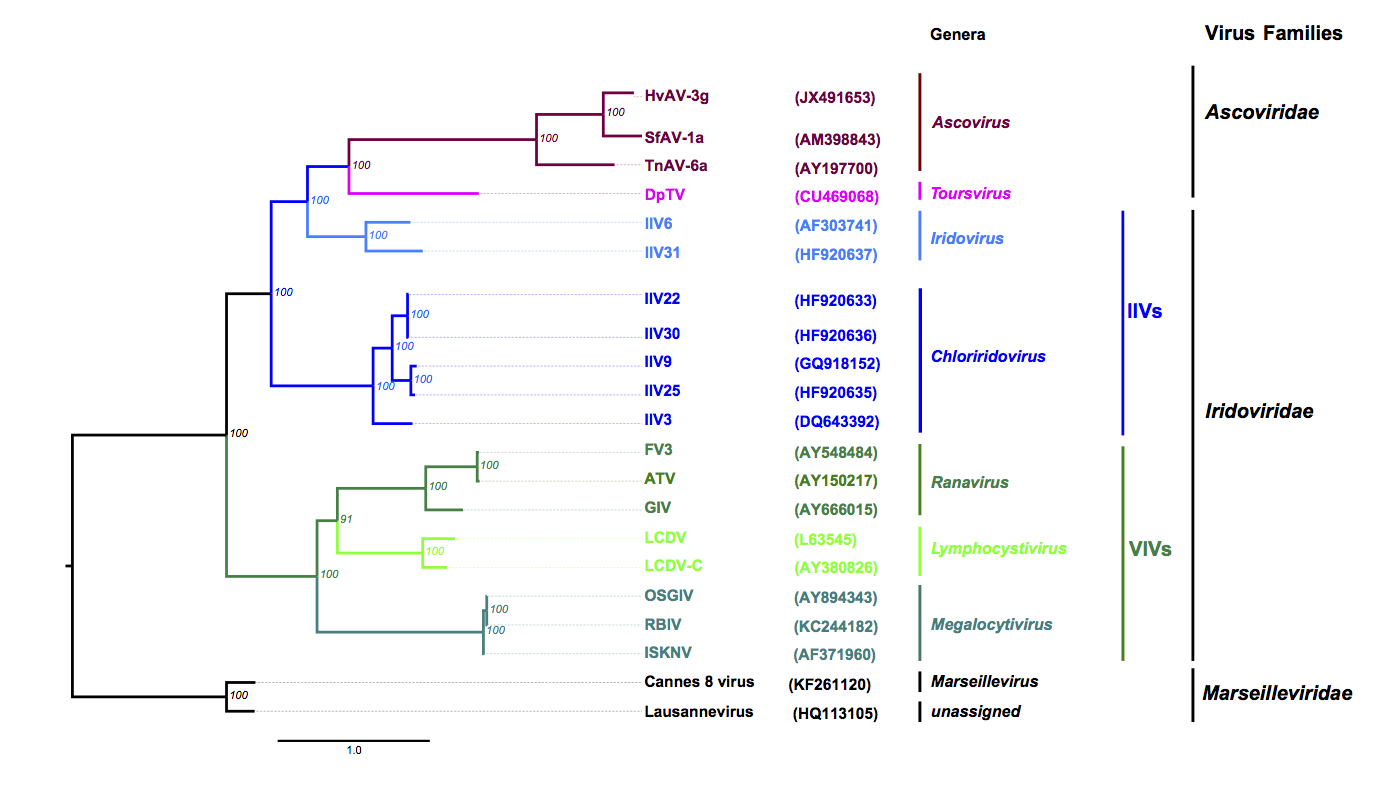 |
| Figure 2 Ascoviridae: Phylogenetic tree obtained with nine core proteins shared by the members of Ascoviridae, Iridoviridae and Marseilleviridae. The tree was calculated using Mafft or Muscle alignments curated with Gblock (parameters were: -t=p -e=-gb1 -b2=N -b3=40 -b4=2 -b5=a -v=120), except for the RNase III orthologues, for which the complete sequence alignment was used. Alignments of the homologues of HvAV-3g ORF1 (DNA polymerase), 11 (DNA-directed RNA polymerase), 15 (DEAD-like helicase), 70 (DNA-directed RNA polymerase II), 74 (hypothetical protein), 81 (hypothetical protein), 85 (serine/threonine protein kinase), 122 (ATPase), 160 (hypothetical protein) were concatenated and trees based on maximum likelihood were calculated with PhyML. Parameters used were WAG (substitution matrix), 0 (proportion of invariable sites), 7 in a, and 5 in f (number of relative substitution rate categories), and F (substitution model). The protein substitution model, the proportion of invariable sites, the number of relative substitution, number of rate categories and substitution model for ML trees were selected and evaluated by ProtTest 3. The best model was chosen on the basis of the Akaike Information Criterion. Numbers in italics at nodes indicate bootstrap values (%) retrieved from 1000 replicates. Branch lengths are proportional to genetic distances. The taxonomic levels from the genera to the families are indicated in the right margin including the non-official grouping of invertebrate iridoviruses (IIVs) and vertebrate iridoviruses (VIVs). Genome accession numbers are shown in brackets. Protein sequences used originate from 8 VIVs including 3 ranaviruses [Ambystoma tigrinum virus (ATV); frog virus 3 (FV3); grouper iridovirus (GIV)], three megalocytiviruses [infectious spleen and kidney necrosis virus (ISKNV); orange-spotted grouper iridovirus (OSGIV); rock bream iridovirus, isolate C1 (TSBIV)] and two lymphocystiviruses [lymphocystis disease virus 1 (LCDV); lymphocystis disease virus, isolate China (LCDV-C)], but also 7 IIVs [IIV3 (Aedes taeniorhynchus iridescent virus), 6 (Chilo iridescent virus), 9 (Wiseana iridescent virus), 22 (Simulium sp. iridescent virus A), 25, 30 (Helicoverpa zea iridescent virus) and 31 (Armadillidium vulgare iridescent virus), 4 ascoviruses (Diadromus pulchellus toursvirus (DpTV), Heliothis virescens ascovirus 3g (HvAV-3g), Trichoplusia ni ascovirus 6a (TnAV-6a), Spodoptera frugiperda ascovirus 1a (SfAV-1a)] and members of two species of Marseilleviridae, the Lausannevirus and the Cannes 8 virus. All these viruses are members of recognized species (except TnAV-6a which is still unclassified) and have their genomes fully sequenced. For more details please refer to (Piegu et al., 2015). Note: The genome of TnAV-2a is not available, therefore, it was not included in the tree. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Phylogenetic analyses using nine core genes shared by the members of Ascoviridae, Iridoviridae and Marseilleviridae revealed that all iridoviruses and members of Ascoviridae have evolved from a common ancestor that they share with members of the family Marseilleviridae (Figure 2. Ascoviridae).

