Family: Filoviridae
Genus: Orthomarburgvirus
Distinguishing features
Two orthomarburgviruses (Marburg virus [MARV] and Ravn virus [RAVV]) are highly lethal human pathogens. Orthomarburgviruses are notable for encoding only a single glycoprotein (including subunits GP1 and GP2) from their GP genes (Feldmann et al., 1992).
Virion
Morphology
Virions are filamentous in shape, but particles can also be branched, circular, U-shaped, or 6 shaped. Spherical forms are rare to absent (Figure 1.Orthomarburgvirus). Virions vary greatly in length (>1 μm) but have a uniform diameter of about 80 nm. Peak infectivity has been associated with particles of about 665 nm in length. Virions are composed of a central core, formed by a helical ribonucleoprotein (RNP) complex, surrounded by a matrix layer and a lipid envelope, derived from the host cell membranes. Spikes about 7 nm in diameter and spaced at intervals of about 10 nm are seen as globular structures on the surface of virions (Ellis et al., 1979a, Ellis et al., 1979b, Geisbert and Jahrling 1995, Ryabchikova and Price 2004, Welsch et al., 2010, Fujita-Fujiharu et al., 2022).
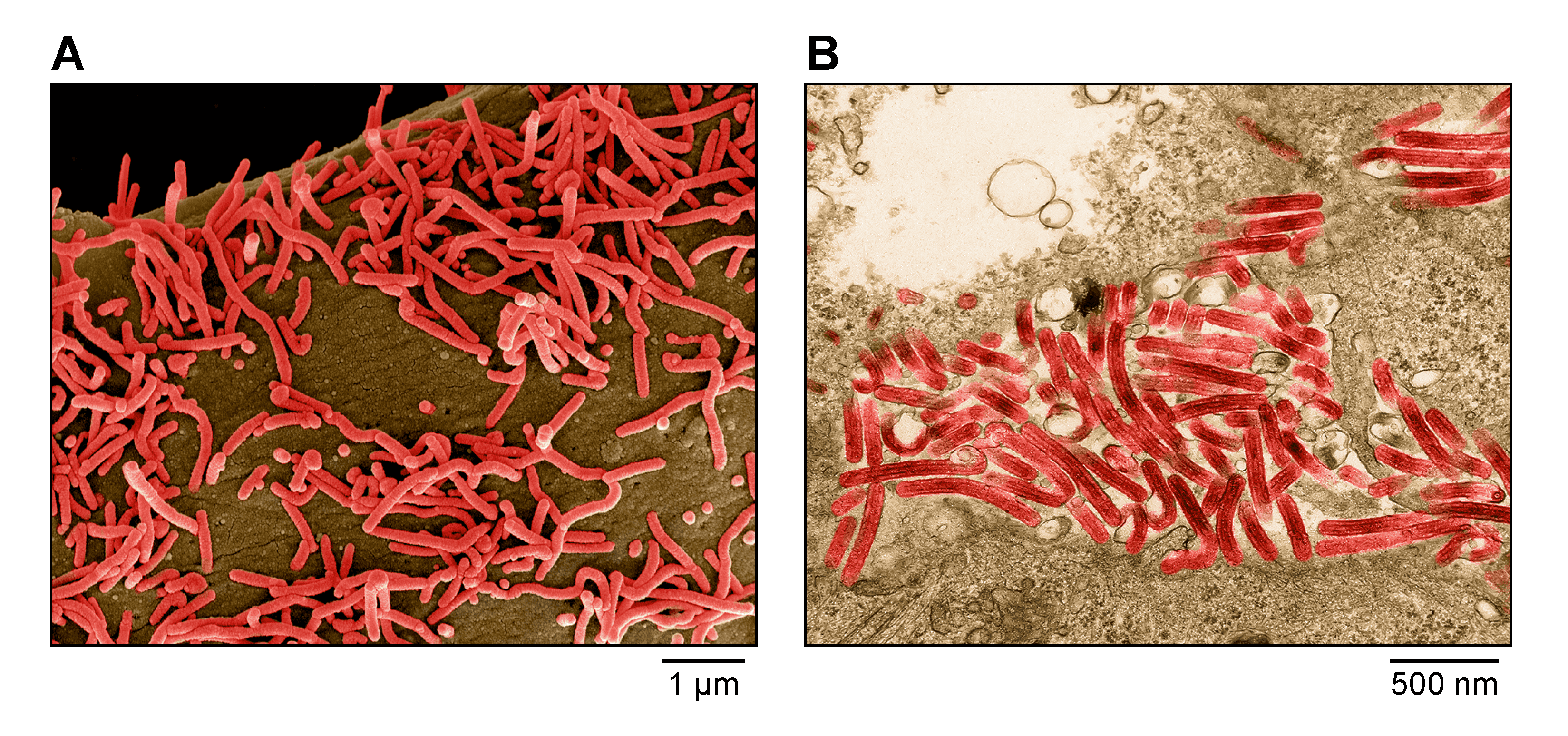 |
| Figure 1. Marburgvirus. A) Scanning electron micrograph of Marburg virus particles (red) budding from an infected grivet (Chlorocebus aethiops (Linnaeus, 1758)) Vero E6 cell. B) Transmission electron micrograph of Marburg virus particles (red) found both as extracellular particles and budding particles from Vero E6 cells. Images are colorized for clarity. Courtesy of John G. Bernbaum and Jiro Wada, NIH/NIAID/DCR/IRF-Frederick, Fort Detrick, MD, USA. |
Physicochemical and physical properties
The buoyant density of Marburg virus (MARV) particles in potassium tartrate is about 1.14 g/mL (Kiley et al., 1988).
Nucleic acid
Orthomarburgvirus genomes are linear non-segmented RNA molecules of negative polarity. Genomic RNAs are about 19.1 kb and are not polyadenylated at their 3′-ends, and there is no evidence of 5′-terminal cap structures or covalently-linked proteins. The Mr of a genomic RNA is about 4.2×106, and the genome represents about 1.1% of the total virion mass (Kiley et al., 1988, Feldmann et al., 1992).
Proteins
Orthomarburgviruses express seven structural proteins (Table 1.Orthomarburgvirus, nucleoprotein [NP], RNP complex-associated protein 24 [VP24], polymerase cofactor [VP35], transcriptional activator [VP30], the matrix protein [VP40], glycoprotein [subunits GP1 and GP2], and large protein [L]), all of which are homologous to those of cuevaviruses, dianloviruses, and orthoebolaviruses. After VP40, the second most abundant structural protein in an orthomarburgvirion is NP, which encapsidates the orthomarburgvirus genome. The least abundant protein is L, which mediates orthomarburgvirus genome replication and transcription (Kiley et al., 1988, Feldmann et al., 1992, Shi et al., 2018). The orthomarburgvirus RNP complex consists of NP, VP24, VP35, VP30, and L. These RNP complexes associate with VP40, which lines the inner side of the virion membrane, and GP1 and GP2, which form globular spikes on the outside of the virion membrane (Mühlberger et al., 1999, Welsch et al., 2010, Kirchdoerfer et al., 2017).
Table 1.Orthomarburgvirus. Location and functions of orthomarburgvirus structural proteins.
| Protein | Encoding gene | Characteristics | Function(s) | Reference |
| Nucleoprotein (NP) | 1 (NP) | RNP complex component; second most abundant protein in infected cells and in virions; consists of two distinct functional modules; homo oligomerizes to form helical polymers; binds to genomic and antigenomic RNA, VP35, VP40, VP30, and VP24; phosphorylated | Nucleocapsid and cellular inclusion body formation; encapsidation of orthomarburgvirus genome and antigenome; genome replication and transcription | (Sanchez et al., 1992, Becker et al., 1994, Becker et al., 1998, Mühlberger et al., 1998, Lötfering et al., 1999, Kolesnikova et al., 2000, Liu et al., 2017, Zhu et al., 2017) |
| Polymerase cofactor (VP35) | 2 (VP35) | RNP complex component; homo oligomer; phosphorylated; binds to double stranded RNA, NP, and L | Replicase transcriptase cofactor; inhibits innate immune response by interfering with IRF3, IRF7, and DDX58; inhibits protein kinase R and PRKRA activity | (Mühlberger et al., 1999, Basler et al., 2000, Basler et al., 2003, Reid et al., 2005, Cárdenas et al., 2006, Haasnoot et al., 2007, Kimberlin et al., 2010, Leung et al., 2010, Biedenkopf et al., 2013, Kirchdoerfer et al., 2015, Nelson et al., 2016) |
| Matrix protein (VP40) | 3 (VP40) | Most abundant protein in infected cells and in virions; consists of two distinct functional modules; homo oligomerizes to form dimers and circular hexamers and octamers; binds single stranded RNA, VP35; hydrophobic; membrane associated; contains one late budding motif; binds NEDD4 and TSG101 | Matrix component; regulation of genome transcription and replication; regulation of virion morphogenesis and egress; inhibits JAK STAT pathway | (Kolesnikova et al., 2002, Kolesnikova et al., 2004a, Kolesnikova et al., 2004b, Kolesnikova et al., 2007, Urata et al., 2007, Urata and Yasuda 2010, Valmas et al., 2010, Valmas and Basler 2011, Kolesnikova et al., 2012, Dolnik et al., 2015, Koehler et al., 2016b, Koehler et al., 2018) |
| Glycoprotein subunits (GP1 and GP2) | 4 (GP) | Type I transmembrane and class I fusion protein; cleaved to GP1 and GP2 subunits that heterodimerize; mature protein is a trimer of GP1 and GP2 heterodimers; inserts into membranes; heavily N- and O glycosylated, acylated, phosphorylated. ADAM17 converts GP1 and GP2 into a soluble form (GP1,2Δ) | Virion adsorption to orthomarburgvirus susceptible cells via cellular attachment factors; determines orthomarburgvirus cell and tissue tropism; induction of virus cell membrane fusion subsequent to endolysosomal binding to NPC1; inhibits innate immune response by interfering with BST2. Function of GP1,2Δ is unknown | (Geyer et al., 1992, Will et al., 1993, Funke et al., 1995, Volchkov et al., 2000, Dolnik et al., 2004, Jouvenet et al., 2009, Sakuma et al., 2009, Carette et al., 2011, Côté et al., 2011, Misasi et al., 2012) |
| Transcriptional activator (VP30) | 5 (VP30) | RNP complex component; hexameric zinc finger protein; binds single-stranded RNA, NP, and L; phosphorylated | Transcription reinitiation at the GP gene start site and potentially antitermination; enhancement of transcription | (Becker et al., 1998, Modrof et al., 2001, Enterlein et al., 2006, Wenigenrath et al., 2010, Albariño et al., 2013, Tigabu et al., 2018, Edwards et al., 2022) |
| RNP complex-associated protein (VP24) | 6 (VP24) | RNP complex component; homo tetramerizes; hydrophobic and membrane associated | Regulation of genome transcription and replication; regulation of virion morphogenesis and egress; targets KEAP1 to activate NFE2L2-induced cytoprotective responses; inhibits both RIGI- and IFIH1-stimulated IFNB and IFNL1 promoter activation | (Bamberg et al., 2005, Lee et al., 2009, Edwards et al., 2014, Page et al., 2014, Zhang et al., 2014, Edwards and Basler 2015, Johnson et al., 2016, Wan et al., 2017, He et al., 2021) |
| Large protein (L) | 7 (L) | RNP complex component; homo dimerizes; binds to genomic and antigenomic RNA, VP35, and VP30; mRNA capping enzyme | Genome replication and mRNA transcription | (Mühlberger et al., 1992, Becker et al., 1998, Mühlberger et al., 1998, Mühlberger et al., 1999, Koehler et al., 2016a, Kirchdoerfer et al., 2017) |
ADAM17, ADAM metallopeptidase domain 17; BST2, bone marrow stromal antigen 2; DDX58, DExD/H-box helicase 58; IFIH1, interferon induced with helicase C domain 1 (formerly MDA5); IFNB, interferon beta; IFNL, interferon lambda; IRF, interferon regulatory factor; JAK-STAT, Janus kinase-signal transducer and activator of transcription; KEAP1, kelch like ECH associated protein 1; NPC1, NPC intracellular cholesterol transporter 1; NEDD4, E3 ubiquitin protein ligase; NFE2L2, nuclear factor, erythroid 2 like 2; PRKRA, protein activator of interferon induced protein kinase EIF2AK2; RNP, ribonucleoprotein; TSG101, tumor susceptibility 101; VP, virus protein
Lipids
The viral envelope is derived from host cell membranes and is considered to have a lipid composition similar to that of the host cell membranes (Bavari et al., 2002). Orthomarburgvirus glycoproteins are acylated (Funke et al., 1995).
Carbohydrates
The glycoproteins of orthomarburgviruses are oligomannosidic contain hybrid-type N glycans and neutral fucosylated bi-, tri-, and tetra-antennary species, most of which carry an additional bisecting N-acetylglucosamine. In addition, the glycoproteins contain O-linked glycans of the neutral mucin type. In contrast to orthoebolavirions, sialylation is rare or absent. Glycans constitute >50% of the GP1 and GP2 total mass (Feldmann et al., 1991, Geyer et al., 1992, Hashiguchi et al., 2015).
Genome organization and replication
The orthomarburgvirus genome has the gene order 3′-NP-VP35-VP40-GP-VP30-VP24-L-5′ (Figure 2.Orthomarburgvirus). The extragenic sequences at the extreme 3′-end (leader) and 5′-end (trailer) of the genome are conserved and partially complementary. Genes are flanked by conserved transcriptional initiation and termination (polyadenylation) sites. Most genes are separated by non conserved intergenic sequences, but two genes overlap. In addition, most genes possess relatively long 3′-end and 5′-end noncoding regions. Similar to dianloviruses, the GP genes of orthomarburgviruses encode only a single protein, GP1,2 (Feldmann et al., 1992, Bukreyev et al., 1995).
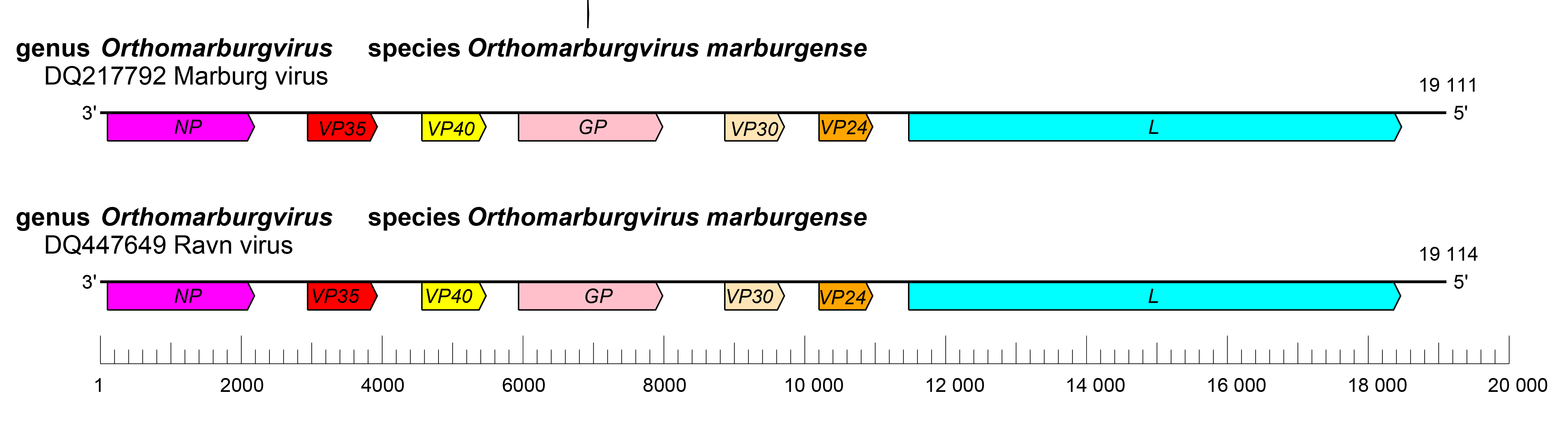 |
| Figure 2. Marburgvirus. Schematic representation of orthomarburgvirus genome organization. Genomes are drawn to scale. |
The replication strategy of orthomarburgviruses is assumed to be highly similar to that of dianloviruses and reminiscent of that of cuevaviruses and orthoebolaviruses (Brauburger et al., 2015, Manhart et al., 2018, Hume and Mühlberger 2019). Orthomarburgvirions enter host cells by endocytosis (Bhattacharyya et al., 2011) after orthomarburgvirus GP1,2 mediates cell surface attachment factor (e.g., C-type lectins, integrins, hepatitis A virus cellular receptor 1 [HAVCR1]) binding and subsequent low pH dependent fusion into endosomes (Davey et al., 2017). Cathepsin L cleavage is required for GP1,2 binding to the endosomal receptor NPC intracellular cholesterol transporter 1 (NPC1), which is also used by cuevaviruses, dianloviruses, and orthoebolaviruses (Carette et al., 2011, Côté et al., 2011, Misasi et al., 2012, Ng et al., 2014). Uncoating is presumed to occur in a manner analogous to that of other mononegaviruses. Orthomarburgvirus transcription and genome replication take place in the cytoplasm and, and, in general, follow the models for other members of the Filoviridae family. Transcription starts at the conserved transcriptional initiation site, and polyadenylation occurs at a stretch of uridine residues within the transcriptional termination site. The 5′-terminal non-coding sequences likely form hairpin like structures in all (capped and polyadenylated) mRNAs. Replication involves the synthesis of full length positive-sense genome copies (antigenomes) (Feldmann et al., 1992, Becker et al., 1998, Mühlberger et al., 1998, Mühlberger et al., 1999, Brauburger et al., 2015). During infection, massive amounts of nucleocapsids accumulate intracellularly and form intracytoplasmic inclusion bodies, which are the sites of orthomarburgvirus transcription, replication, and nucleocapsid assembly. Mature nucleocapsids are transported for envelopment to the plasma membrane, where budding occurs in a VP40-mediated process (Kolesnikova et al., 2002, Kolesnikova et al., 2004a, Kolesnikova et al., 2004b, Kolesnikova et al., 2007, Kolesnikova et al., 2012, Schudt et al., 2013, Dolnik et al., 2015, Takamatsu et al., 2019) (Figure 3.Filoviridae).
Biology
Orthomarburgviruses are endemic in Eastern Africa (MARV, RAVV), Middle Africa (MARV), Southern Africa (MARV), and Western Africa (MARV). Egyptian rousettes (Rousettus aegyptiacus (Geoffroy, 1810)) are naturally infected by both orthomarburgviruses. The route of initial human infection is unknown (Towner et al., 2009, Amman et al., 2012, Amman et al., 2014, Amman et al., 2017, Pawęska et al., 2018). The major route of human-to-human transmission of orthomarburgviruses requires direct contact with blood, bodily fluids, or injured skin. Both orthomarburgviruses are highly lethal human pathogens (Kuhn et al., 2020). In the laboratory, rodents (laboratory mice, domesticated guinea pigs, golden hamsters) and nonhuman primates (common marmosets, crab-eating macaques, grivets, hamadryas baboons, rhesus monkeys, squirrel monkeys) can be infected experimentally with various orthomarburgviruses, but lethal infection of rodents requires sequential adaptation (Kuhn 2020, St Claire et al., 2017, Siragam et al., 2018). In contrast to orthoebolaviruses, orthomarburgviruses do not cause disease in domestic ferrets (Cross et al., 2018, Wong et al., 2018).
Antigenicity
The antigenicity of orthomarburgvirions is primarily due to their glycoproteins. Several anti GP1,2 monoclonal antibodies are specific for orthomarburgviruses. The ability of a monoclonal antibody to neutralize orthomarburgvirus infection in vitro may not necessarily be predictive of protective efficacy in vivo; conversely, antibodies that are non-neutralizing in vitro may be protective in vivo. There is only limited antigenic relatedness between the virions of orthomarburgviruses and other filovirids (Flyak et al., 2015, Hashiguchi et al., 2015, Froude et al., 2017, Mire et al., 2017, Sangha et al., 2017, King et al., 2018, Marzi et al., 2018).
Species demarcation criteria
The genus currently includes only a single species.
Phylogenetic relationships across the genus have been established from maximum likelihood trees generated using coding-complete or complete genome sequences (Figure 4.Filoviridae) or by phylogenetic analysis of RNA-directed RNA polymerase (RdRP) sequences (Wolf et al., 2018). MARV and RAVV are relatively distinct in genomic sequence similarity analyses but occupy the same ecological niche and cause the same disease in humans and nonhuman primates. Hence, they are currently considered distinct viruses belonging to the same species.

