Family: Kolmioviridae
Jens H. Kuhn, Artem Babaian, Laura M. Bergner, Paul Dény, Dieter Glebe, Masayuki Horie (堀江真行), Eugene V. Koonin, Mart Krupovic, Sofia Paraskevopoulou (Σοφία Παρασκευοπούλου), Marcos de la Peña, Teemu Smura, and Jussi Hepojoki
The citation for this ICTV Report chapter is the summary published as Kuhn et al. (2024):
ICTV Virus Taxonomy Profile: Kolmioviridae, Journal of General Virology 2024, (in press)
Corresponding author: Jussi Hepojoki (jussi.hepojoki@helsinki.fi)
Edited by: Holly Hughes and Evelien Adriaenssens
Posted: January 2024
Summary
Kolmioviridae is a family for negative-sense RNA viruses with circular viroid-like genomes of about 1.5–1.7 kb (Table 1.Kolmioviridae). The family includes eight genera (Daazvirus, Dagazvirus, Daletvirus, Dalvirus, Deevirus, Deltavirus, Dobrovirus, and Thurisazvirus). Kolmiovirids are maintained in and/or transmitted by amphibian, birds, fish, insects, mammals, and reptiles. Deltaviruses can cause severe hepatitis and possibly other diseases in humans. Kolmiovirids encode at least one protein (delta antigen; DAg). They use host-cell DNA-directed RNA polymerase II and contain ribozymes in their genome (negative sense) and antigenome (positive sense) to achieve replication. They require evolutionarily unrelated helper viruses to provide envelopes with incorporated helper virus surface proteins for infectious particle formation.
Table 1.Kolmioviridae. Characteristics of members of the family Kolmioviridae.
| Characteristic | Description* |
| Example | hepatitis D virus 1 (AF104263), species Deltavirus italiense, genus Deltavirus |
| Virion | Spherical virions (36–43 nm in diameter) with an outer envelope containing envelope proteins derived from a helper virus and an inner ribonucleoprotein (RNP) consisting of genomic RNA and a nucleoprotein (delta antigen; DAg) that may occur in two isoforms |
| Genome | Non-segmented, ribozyme-containing, negative-sense, covalently closed circular RNA (cccRNA) of about 1.5–1.7 kb that forms a rod-like structure through a high degree of self-complementarity |
| Replication | RNA-directed RNA synthesis by host-cell DNA-directed RNA polymerase II through a double rolling circle mechanism, and autocatalytic cleavage/ligation via encoded genomic and antigenomic ribozymes and re-cyclization in the nucleus |
| Translation | mRNA-based translation of DAg (and in some cases including an isoform thereof) |
| Host range | Kolmiovirids infect amphibians, birds, fish, insects, mammals, and reptiles |
| Taxonomy | Realm Ribozyviria. The family includes eight genera and 15 species |
* mostly based on experiments with mammalian deltaviruses
Viruses assigned to each of the eight genera form a monophyletic clade based on phylogenetic analysis of delta antigen (DAg) homologs. Viruses from all eight genera share at least two of the following characteristics: (i) enveloped spherical virions; (ii) circular negative-sense RNA genome containing ribozymes and encoding a DAg homolog; (iii) requiring an unrelated helper virus for assembly of infectious virions.
Amphibian Hosts
Genus Daazvirus. Viruses in this genus infect salamandrid amphibians (newts) (Chang et al., 2019).
Genus Dobrovirus. Viruses in this genus infect bufonid amphibians (toads) (Chang et al., 2019).
Avian Hosts
Genus Dalvirus. Viruses in this genus infect anatid birds (ducks and teals) (Wille et al., 2018).
Insect Hosts
Genus Dagazvirus. Viruses in this genus infect rhinotermitid insects (termites) (Chang et al., 2019).
Piscine Hosts
Genus Deevirus. Viruses in this genus infect unspecified fish (Chang et al., 2019).
Mammalian Hosts
Genus Deltavirus. Viruses in this genus infect humans and possibly dasyurid marsupials (dunnarts and Tasmanian devils) (Rizzetto et al., 1980, Rizzetto 2015, Harvey et al., 2023).
Genus Thurisazvirus. Viruses in this genus infect echimyid rodents (spiny-rats) and possibly phyllostomid bats (vampire bats) (Paraskevopoulou et al., 2020, Bergner et al., 2021).
Reptilian Hosts
Genus Daletvirus. Viruses in this genus infect boid reptiles (snakes) (Hetzel et al., 2019).
Virion
Morphology
Only known for members of the genus Deltavirus (see Deltavirus genus page).
Physicochemical and physical properties
Only known for members of the genus Deltavirus (see Deltavirus genus page).
Nucleic acid
Kolmiovirids have a nonsegmented, viroid-like, ribozyme-containing, negative-sense, covalently closed circular RNA (cccRNA) genome of about 1.5–1.7 kb (Figure 1 Kolmioviridae) that forms a rod-like structure through its high degree of self-complementarity. Further information is only known for members of the genus Deltavirus (see Deltavirus genus page).
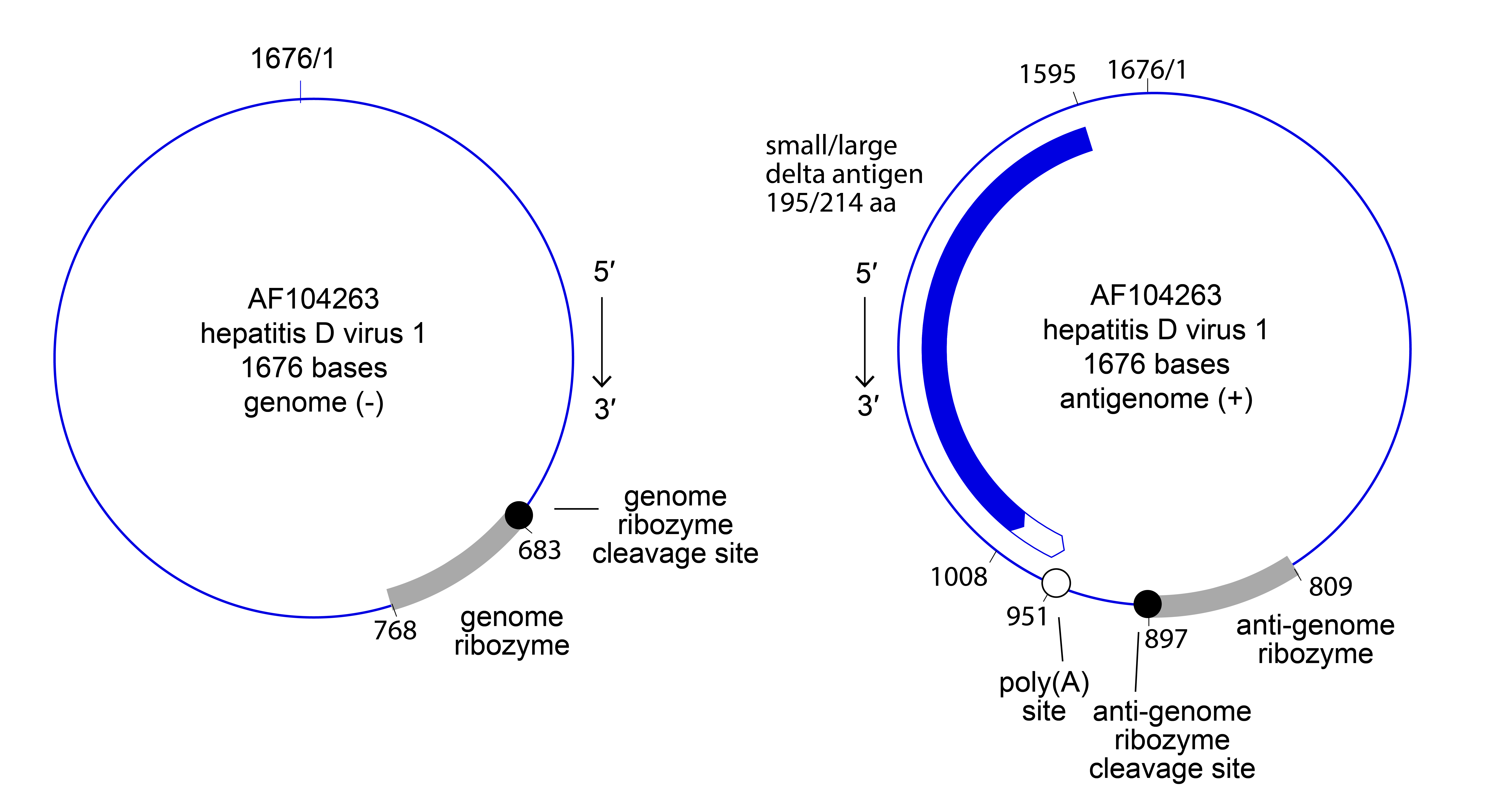 |
| Figure 1 Kolmioviridae. Genome and antigenome organization of the kolmiovirid hepatitis D virus 1. Nucleotide positions on the antigenome are numbered according to those for the (complementary) genome. |
Proteins
Kolmiovirids encode one protein, delta antigen (DAg). Some kolmiovirids (deltaviruses) express two isoforms of DAg. Further information is only known for members of the genus Deltavirus (see Deltavirus genus page).
Lipids
Not reported but are likely derived from host-cell membranes via helper virus envelope protein-assisted budding and therefore likely composed of phospholipids, glycolipids, fatty acids, and sterols.
Carbohydrates
Only known for members of the genus Deltavirus (see Deltavirus genus page).
Genome organization and replication
Kolmiovirids enter host cells through engagement of cell-surface receptors using envelope proteins provided by their helper viruses; consequently, the host and tissue tropism of a particular kolmiovirid reflects that of its helper virus. Fusion of the kolmiovirion membrane with host-cell membranes releases the ribonucleoprotein (RNP) into the cytosol, from where it migrates to the cell nucleus. Kolmiovirid RNA-directed RNA (genome and antigenome) synthesis is mediated by host-cell DNA-directed RNA polymerase II through a double rolling circle mechanism; genome- and antigenome-encoded ribozymes catalyze autocatalytic cleavage of concatenated progeny genomes and ligation/recyclization in the nucleus. DAg expression induces packaging of the kolmiovirid RNPs into progeny virions, which bud in a process mediated by helper virus proteins (Figure 2 Kolmioviridae). Specifics are only known for members of the genus Deltavirus (see Deltavirus genus page), and helper viruses have only been identified for deltaviruses and daletviruses.
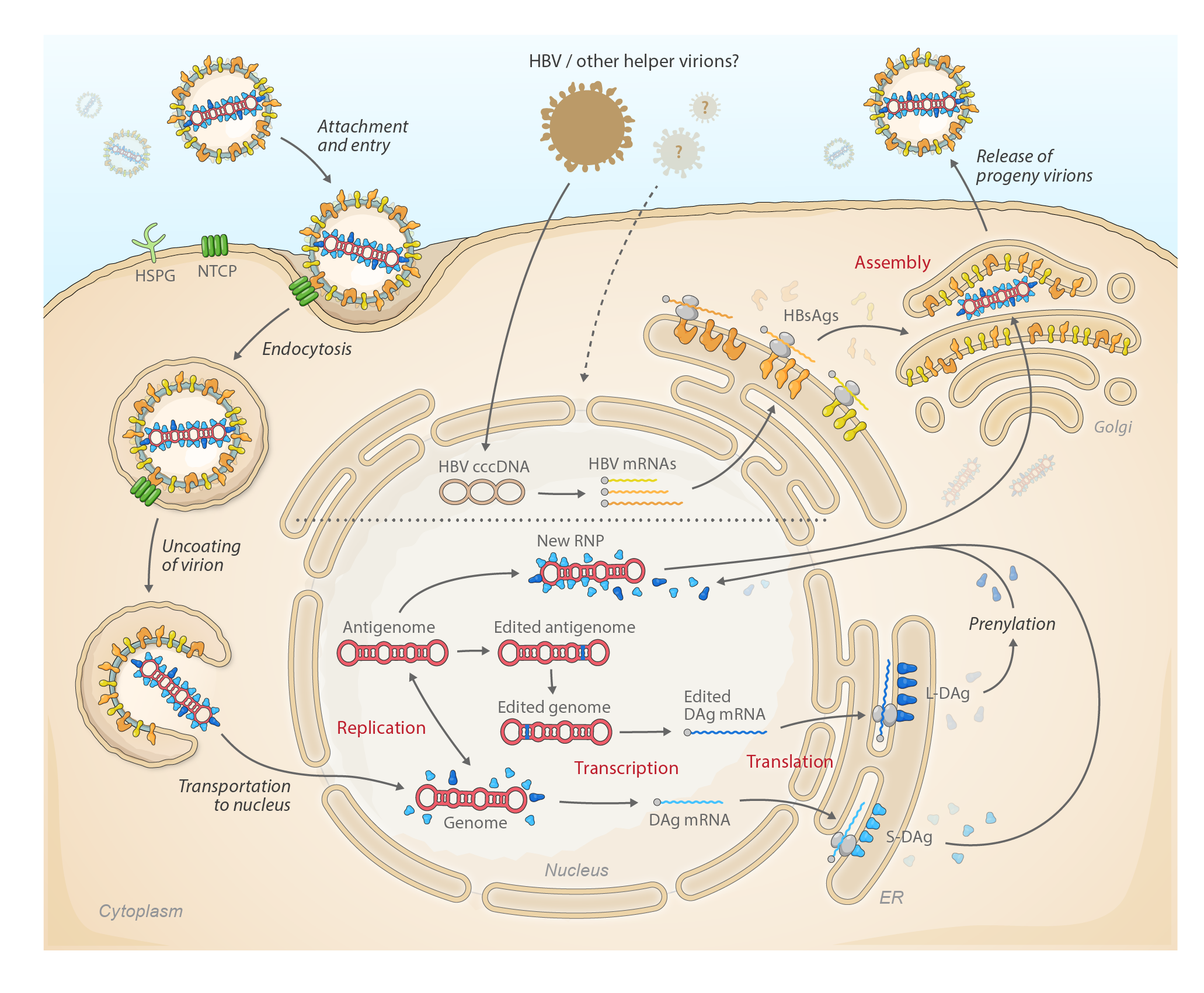 |
| Figure 2 Kolmioviridae. Lifecycle of kolmiovirids. |
Biology
Kolmiovirids have been found in bufonid and salamandrid amphibians, anatid birds, unspecified fish, rhinotermitid insects, boid reptiles, and humans and echimyid rodents (Wille et al., 2018, Chang et al., 2019, Hetzel et al., 2019, Lucifora and Delphin 2020, Paraskevopoulou et al., 2020, Bergner et al., 2021, Asselah and Rizzetto 2023). Unclassified, putative kolmiovirids have been discovered in common vampire bats (phyllostomid Desmodus rotundus (Geoffroy, 1810)), groundhogs (sciurid Marmota monax (Linnaeus, 1758), white-tailed deer (cervid Odocoileus virginianus (Zimmermann, 1780)), fat-tailed dunnarts (dasyurid Sminthopsis crassicaudata (Gould, 1844)), Tasmanian devils (dasyurid Sarcophilus harrisii (Boitard, 1841)), white-rumped munias (estrildid Lonchura striata (Linnaeus, 1766)), yellow-bellied tits (parid Pardaliparus venustulus (R. Swinhoe, 1870)), and Australian zebra finches (estrildid Taeniopygia castanotis (Gould, 1837)), and in in fungus-growing termites (termitid Odontotermes wallonensis Wasmann, 1902) (Bergner et al., 2021, Iwamoto et al., 2021, Litov et al., 2022, Harvey et al., 2023).
Because the infectivity of kolmiovirids is believed to be dependent on evolutionarily unrelated helper viruses, such as viruses of the families Hepadnaviridae and Arenaviridae, host tropism of particular kolmiovirids is thought to be primarily determined by the distribution of the respective helper virus. Only deltaviruses are known to cause diseases (see Deltavirus genus page).
Antigenicity
Systematic antigenicity studies have only been reported for deltaviruses (see Deltavirus genus page). As kolmiovirids only encode DAg or DAg isoforms, presence of antibodies to DAg is indicative of current or past infections (Rizzetto 2015).
Derivation of names
actinopterygii: from the fish host class Actinopterygii
anatis: from the bird host genus Anas
boae: from the snake host genus Boa
bufonis: from the amphibian host genus Bufo
cameroonense: from Cameroon
carense: from Central African Republic
cynopis: from the amphibian host genus Cynops
Daazvirus: from the Gothic letter Daaz (̳) – a successor of the Greek letter Δ spelled out in genus Deltavirus
Dagazvirus: from the Runic letter Dagaz (ᛞ), a possible descendant of the Old Italic D – a successor of the Greek letter Δ spelled out in genus Deltavirus
Daletvirus: from the Hebrew letter 'Dālet (ד) – a predecessor of the Greek letter Δ spelled out in genus Deltavirus
Dalvirus: from the Arabic letter Dal (د) (pronounced similarly to the English letter “d”) – a successor of the Greek letter Δ spelled out in genus Deltavirus
Deevirus: from the English letter Dee (D) – a successor of the Greek letter Δ spelled out in genus Deltavirus
Deltavirus: from δ (delta) antigen (δAg) originally identified in HBV-infected tissue but unrelated to previously described HBV antigens (Rizzetto et al., 1977).
Dobrovirus: from the Early Cyrillic letter Dobro (Д) – a successor of the Greek letter Δ spelled out in genus Deltavirus
italiense: from Italy
japanense: from Japan
Kolmioviridae: from the Finnish kolmio, meaning “triangle” – a tongue-in-cheek reference to the the Greek letter Δ spelled out in genus Deltavirus
myis: from the rodent host genus Mus
peruense: from Peru
schedorhinotermitis: from the blattodean host genus Schedorhinotermes
senegalense: from Senegal
taiwanense: from Taiwan
Thurisazvirus: from the Runic letter Thurisaz (ᚦ), a possible descendant of the Old Italic D – a successor of the Greek letter Δ spelled out in genus Deltavirus
togense: from Toga
Genus demarcation criteria
Members of different genera encode DAg with ≤60% amino acid sequence similarity.
Relationships within the family
Relationships across the family have been estimated via Bayesian phylogeny using complete small antigen amino acid sequences (Figure 3 Kolmioviridae).
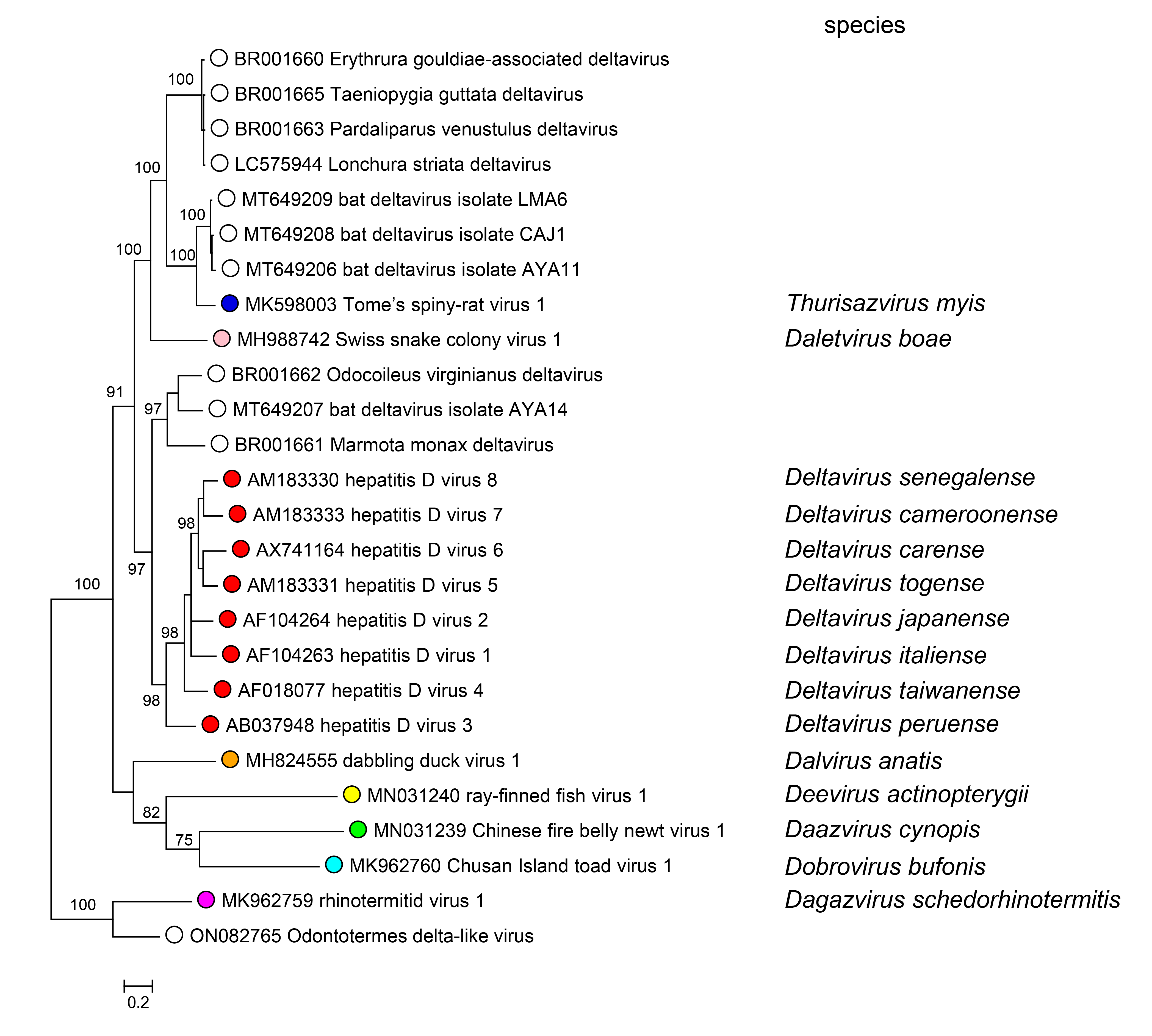 |
| Figure 3 Kolmioviridae. Maximum clade credibility tree based on a (small) delta antigen (DAg) amino acid alignment from representatives of all available kolmiovirids. Posterior probabilities are shown for all nodes. Tree tips correspond to GenBank accession numbers. The phylogenetic tree was constructed using the Bayesian Markov chain Monte Carlo (MCMC) method, implemented in MrBayes version 3.2 (Ronquist et al., 2012) with two independent runs and four chains per run, and the JTT model of substitution. The analysis was run for 5 million states and sampled every 5,000 steps. The average standard deviation of split frequencies was 0.010. |
Relationships with other taxa
Kolmiovirids are unrelated to all other negative-sense RNA viruses, which are evolutionarily connected through their encoded RNA-directed RNA polymerases (RdRPs). Several kolmiovirid features, such as genome structure, RNA transcription using host-cell DNA-directed RNA polymerase II, the presence of autocatalytic RNA sites, and RNA-to-RNA double rolling circle replication are similar to those of viroids, certain virusoids, certain helper virus-dependent viroid-like satellite RNAs (Flores et al., 2012, Taylor 2015, de la Peña et al., 2021, Forgia et al., 2021, Edgar et al., 2022, Forgia et al., 2023), and recently discovered mobile genetic elements (e.g., “epsiloviruses”, “zetaviruses”), some of which have an organization reminiscent of kolmiovirids but also encode orthornaviraen RdRPs (“Ambiviricota”). The evolutionary origins of kolmiovirids and viroids and their evolutionary relationships are uncertain (Elena et al., 1991, Jenkins et al., 2000).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| bat deltavirus DesRot/Peru/AYA14_DrDV-A | MT649207 | (Bergner et al., 2021) | |
| Marmota monax deltavirus | BR001661 | (Iwamoto et al., 2021) | |
| Odocoileus virginianus deltavirus | BR001662 | (Iwamoto et al., 2021) | |
| Taeniopygia guttata deltavirus | BR001665 | (Iwamoto et al., 2021) | |
| Lonchura striata deltavirus | LC575944 | (Iwamoto et al., 2021) | |
| Pardaliparus venustulus deltavirus | BR001663 | (Iwamoto et al., 2021) | |
| Cát Tiên Odontotermes delta-like virus | ON082765 | (Litov et al., 2022) |
Virus names and virus abbreviations are not official ICTV designations.
*Genome sequence incomplete.
Additional unclassified kolmiovirids that are probable members of established genera are listed under individual genus descriptions.

