Family: Nudiviridae
Monique M. van Oers, Annie Bézier, Elisabeth A. Herniou, Johannes A. Jehle, Kelly S. Bateman, Madoka Nakai, Robert L. Harrison
The citation for this ICTV Report chapter is the summary published as Harrison et al., (2020):
ICTV Virus Taxonomy Profile: Nudiviridae, Journal of General Virology, 101, 3–4.
Corresponding author: Monique M. van Oers (monique.vanoers@wur.nl)
Edited by: Balázs Harrach, Stuart G. Siddell and Arvind Varsani
Posted: December 2019, updated May 2022, May 2023
PDF: ICTV_Nudiviridae.pdf (2019 version)
Summary
Members of the family Nudiviridae are large dsDNA viruses with distinctive rod-shaped nucleocapsids and circular genomes of 96 to 232 kbp. Nudiviruses have been identified from a diverse range of insects and crustaceans. Although nudiviruses resemble baculoviruses and these families share a number of core genes, they are part of a separate, distinct lineage of the order Lefavirales.
Table 1.Nudiviridae Characteristics of members of the family Nudiviridae
|
Characteristic |
Description |
|
Example |
Oryctes rhinoceros nudivirus Ma07 (EU747721), species Alphanudivirus oryrhinocerotis, genus Alphanudivirus |
|
Virion |
Enveloped, rod-shaped or ellipsoidal, compact (approximately 100 nm × 200 nm) or elongated (approximately 81 nm × 415 nm) |
|
Genome |
A single covalently-closed circular dsDNA molecule of 96–232 kbp encoding 89–155 proteins |
|
Replication |
Nuclear, with nucleocapsids assembled and enveloped within the nucleus |
|
Translation |
From mRNAs transcribed from viral DNA |
|
Host range |
Immature and adult stages of insects and crustaceans |
|
Taxonomy |
Class Naldaviricetes, order Lefavirales. The family has four genera with thirteen species |
Virion
Morphology
Virions consist of bacilliform nucleocapsids surrounded by an envelope. The virions can be relatively compact (100 nm × 200 nm; (Payne et al., 1977) or elongated (81 × 415 nm) (Hamm et al., 1996). In some cases, a characteristic bulb shaped protuberance of the envelope is observed at one end of the virion (Stentiford et al., 2004, Bateman et al., 2021). Tail-like appendages extending from the nucleocapsid are sometimes present (Payne et al., 1977, Hamm et al., 1996). Virions assembled into occlusion bodies have been noted in nudivirus-infected hosts in some instances (Smith 1956, Lightner and Redman 1981, Huger 1991, Raina et al., 2000, Bézier et al., 2015).
Physicochemical and physical properties
Virions have a buoyant density of 1.18–1.23 g/mL in sucrose gradients (Payne 1974, Huger 1985) and 1.28 g/mL in CsCl gradients (Payne et al., 1977). The envelope surrounding the virion is removed upon treatment with the detergent NP-40, and the remaining nucleocapsids possess a density of 1.47 g/mL in CsCl gradients (Payne et al., 1977).
Nucleic acid
Nudivirus DNA consists of a supercoiled, circular double-stranded molecule (Huang et al., 1982) and genomes of 96 to 232 kbp have been described.
Proteins
Annotations of nudivirus genomes indicate that nudiviruses encode an estimated 89 to 155 proteins. Protein gels of purified virions have distinguished 26–28 structural proteins (Burand et al., 1983, Crawford and Sheehan 1985), and proteomic analysis determined that the occlusion bodies of a nudivirus from the crane fly, Tipula oleracea, consist of 48–52 virus-encoded proteins (Bézier et al., 2017). Despite earlier claims, the gene(s) encoding occlusion body matrix protein(s) of nudiviruses have not been identified (Wang et al., 2011, Bézier et al., 2015). The nudivirus occlusion body matrix most likely consists of a protein unrelated to baculovirus polyhedrin (Chaivisuthangkura et al., 2008). Baculovirus homologs of several nudivirus genes encode structural components of baculovirus virions, such as the major capsid protein VP39 and the envelope protein P74 and other per os infectivity factors. Genes for viral replication, such as DNA polymerase, and RNA polymerase components LEF-8, LEF-9, and P47 are also conserved between these two virus families.
Lipids
Lipids are likely present in the envelope but have not been characterized.
Carbohydrates
Carbohydrates are present in nudiviral glycoproteins (Burand et al., 1983).
Genome organization and replication
ORFs are distributed throughout the genome, with approximately equal numbers of the ORFs occurring in either orientation (Figure 1.Nudiviridae) (Cheng et al., 2002, Wang et al., 2007b, Wang et al., 2011, Burand et al., 2012, Yang et al., 2014, Bézier et al., 2015, Hill and Unckless 2018, Holt et al., 2019). The order of homologs shared among nudivirus genomes is poorly conserved. Twenty-eight core genes have been identified as being present in all nudivirus genomes, including twenty-one homologs of baculovirus core genes (Table 2.Nudiviridae) (Bézier et al., 2015). In addition, intergenic regions of direct or tandem repeated sequences are present. TATA box and promoter motifs, including the baculovirus late promoter motif TAAG, have been identified upstream of a number of nudivirus ORFs, (Cheng et al., 2002, Wang et al., 2007c, Bézier et al., 2015).
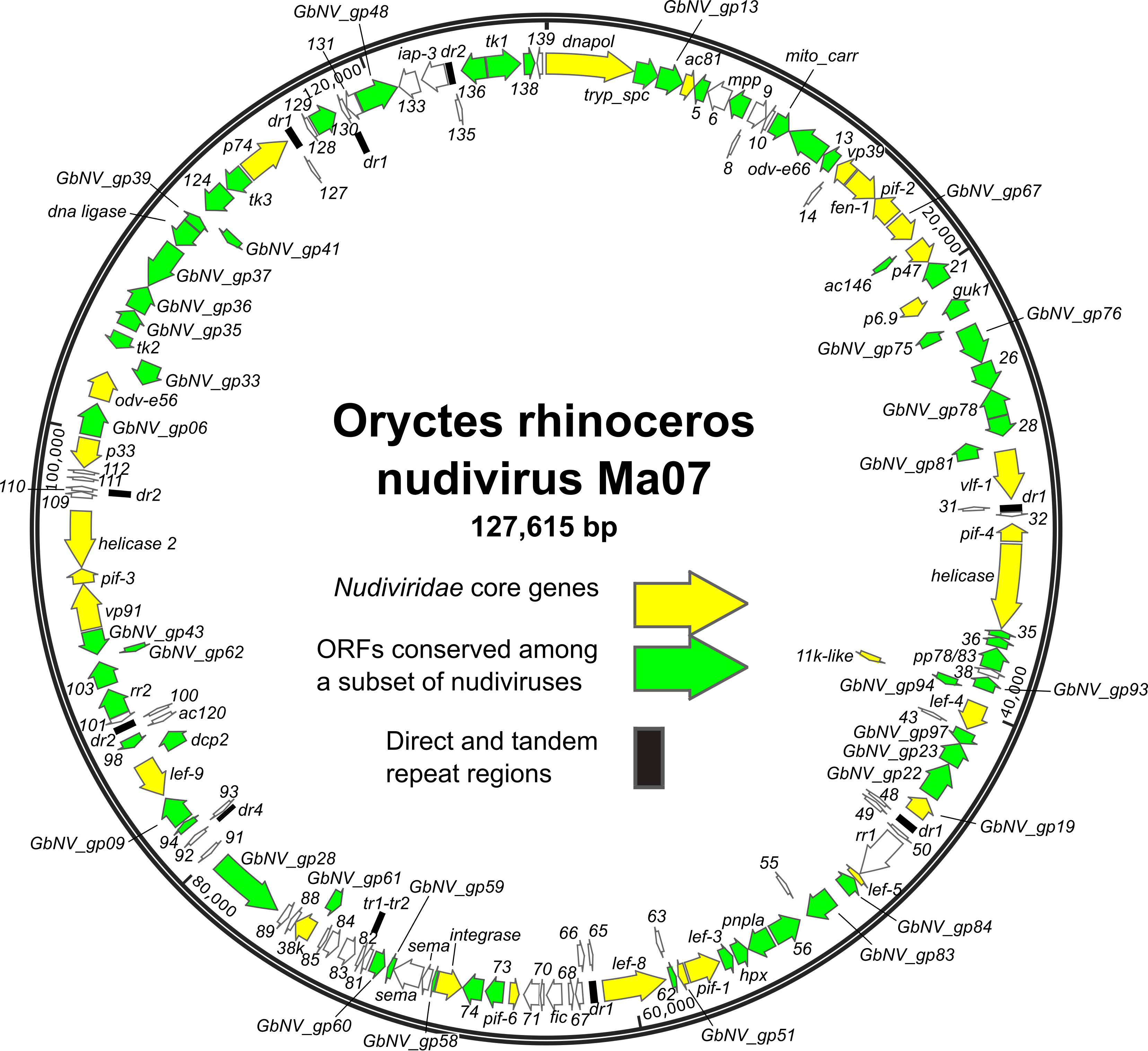 |
|
Figure 1.Nudiviridae. Genome map of Oryctes rhinoceros nudivirus isolate Ma07, the exemplar isolate of the species Alphanudivirus oryrhinocerotis. The map is illustrated with locations and orientations of annotated ORFs (arrows). ORFs corresponding to the core genes of members of the family Nudiviridae, and ORFs conserved among subsets of nudiviruses are indicated. Locations of direct (dr) and tandem (tr) repeat sequence regions are also shown. |
Nudivirus infection of cells results in cytopathology characterized by cellular rounding and nuclear hypertrophy (Huger 1966, Crawford and Sheehan 1985, Burand and Lu 1997, Rallis and Burand 2002a). Virus-specific transcripts and infected cell-specific proteins appear at different times during infection (Burand et al., 1983, Crawford and Sheehan 1985, Chao et al., 1992), suggesting that there is temporal regulation of viral gene expression. Nuclear envelopment has been observed earlier (Kelly et al., 1981, Crawford and Sheehan 1985) and accumulating progeny virions have been described in paracrystalline arrays, within the nucleus or within cytoplasmic vesicles (Huger 1966, Hamm et al., 1996, Burand and Lu 1997, Rallis and Burand 2002a). An in-depth EM study of Oryctes rhinoceros nudivirus (OrNV) infections in cultured cells (Velamoor et al., 2020) revealed that the nuclear virogenic stroma is associated with viral replication and the nucleus becomes filled with tubulars structures surrounded by virions. Invaginations of the inner and outer nuclear membrane into the nucleus are also frequently seen. These observations lead to the hypothesis that the tubular structures are involved in DNA packaging, while the membrane protrusions provide material for envelopment of nucleocapsids. Furthermore, packaging and envelopment seem to be simultaneously coordinated nuclear process.
There is evidence that the nuclear membrane intrusions also play a role in the transport of enveloped virions over the double nuclear membrane (Velamoor et al., 2020). Late in infection the endoplasmic reticulum and Golgi apparatus disintegrate and numerous cytoplasmic vesicles with and without virions are being formed. Shedding of virus particles from the cell surface is observed well before cell lysis, both as virions and as multilayer vesicles. Finally cells loose complete integrity, thereby releasing virions together with nuclear and cytoplasmic material (Burand and Lu 1997, McIntosh et al., 2007, Velamoor et al., 2020).
Table 2.Nudiviridae. Core genes of members of the family Nudiviridae.
The number of recognized core genes was reduced from 32 to 28 after the sequencing of the Crangon crangon nudivirus (Bateman et al., 2021).
|
Oryctes rhinoceros nudivirus (OrNV) ORF |
Gene name |
Inferred activity or function of encoded protein |
|
ORF1a |
dnapol |
DNA polymerase |
|
ORF4a |
ac81 |
Nucleocapsid envelopment |
|
ORF15a |
vp39 |
Major capsid protein |
|
ORF16 |
fen-1 |
FEN-1/FLAP endonuclease |
|
ORF17a |
pif-2 |
Per os infectivity factor |
|
ORF18 |
GbNV_gp67-like |
Unknown |
|
ORF20a |
p47 |
RNA polymerase subunit |
|
ORF22a, b |
p6.9 |
Nucleocapsid packaging/assembly |
|
ORF30a |
vlf-1 |
Very late gene expression factor |
|
ORF33a |
pif-4 |
Per os infectivity factor |
|
ORF34a |
helicase |
DNA helicase |
|
ORF41 |
11K-like |
Viral particle component |
|
ORF42a |
lef-4 |
RNA polymerase subunit |
|
ORF47 |
GbNV_gp19-like |
Occlusion body component |
|
ORF52a |
lef-5 |
Transcription initiation factor |
|
ORF60a |
pif-1 |
Per os infectivity factor |
|
ORF61 |
GbNV_gp51-like |
Viral particle component |
|
ORF64a |
lef-8 |
RNA polymerase subunit |
|
ORF72a |
pif-6 |
Per os infectivity factor |
|
ORF75 |
integrase |
DNA processing |
|
ORF87a |
38K |
Nucleocapsid protein, viral phosphatase |
|
ORF96a |
lef-9 |
RNA polymerase subunit |
|
ORF106a |
vp91/p95 |
Nucleocapsid protein |
|
ORF107a |
pif-3 |
Per os infectivity factor |
|
ORF108 |
helicase-2 |
DNA helicase |
|
ORF113a |
p33 |
Sulfhydryl oxidase |
|
ORF115a |
odv-e56/pif-5 |
Per os infectivity factor |
|
ORF126a |
p74/pif-0 |
Per os infectivity factor |
a Also conserved among members of the family Baculoviridae
b The p6.9 homolog of OrNV and related alphanudiviruses from drosophilid hosts is a fusion of a portion of the Gryllus bimaculatus nudivirus (GbNV) p6.9 (ORF73) and part of the upstream GbNV ORF72. In other nudiviruses, p6.9 exists as an independent ORF.
Biology
Nudiviruses have been isolated from insects of the orders Coleoptera, Diptera, Lepidoptera, and Orthoptera, and from crustaceans of the orders Amphipoda and Decapoda, although some of these viruses are unclassified. Infections of both adult and immature stages of the host can occur. Transmission occurs horizontally and is initiated by oral ingestion of virions followed by infection of the midgut (Zelazny 1976, Huger 1985, Hamm et al., 1996, Raina and Lupiani 2006, Bézier et al., 2015). In addition, horizontal transmission occurring via sexual contact between adults has been documented for some nudiviruses, as has vertical transmission from infected adults to progeny (Hamm et al., 1996, Burand et al., 2004). Viral DNA by itself can initiate an infection when transfected into cultured cells (McIntosh et al., 2007). The outcome of infection ranges from cell lysis and ultimate mortality of the infected host to establishment of a persistent, asymptomatic infection.
Genus demarcation criteria
Because relatively low numbers of nudiviruses have been identified and studied, criteria for the demarcation of nudivirus genera are not well-defined. Currently, nudivirus genera can be broadly distinguished by molecular phylogeny based on core genes and differences in genome characteristics (size, GC and gene content), and virion dimensions, host species, tissue tropism and mode of transmission. Analyses that include sequences of unclassified nudiviruses suggest the possibility that pairwise amino acid distances of selected core genes and the presence or absence of core gene synteny may be used in the future to develop demarcation criteria for nudivirus genera with more precision. Similar analyses have led to the discrimination of four, instead of the original two, genera of nudiviruses.
Derivation of names
Alphanudivirus, Betanudivirus, Gammanudivirus, Deltanudivirus: from the Greek letters α, β, γ and δ, the first four letters of the Greek alphabet
Nudiviridae: from the Latin “nudus”, meaning “naked”, referring to virions that are not occluded in a baculovirus-like occlusion protein matrix. This prefix is taken from “Nudibaculovirinae”, the name of the former subfamily of Baculoviridae into which nudiviruses had been previously classified in the 5th Report of the ICTV. At that time, it was not known that some nudiviruses do form occlusion bodies.
Nudivirus species were changed to binomial format in 2023 following ratification of taxonomy proposal 2022.003D.A.Lefavirales_106rensp.docx. The common names for nudivirus isolates have not been changed.
Relationships within the family
Nudiviruses in the genera Alphanudivirus, Betanudivirus, Gammanudivirus and Deltanudivirus form distinct phylogenetic groupings, based upon analysis of core gene amino acid sequences (Figure 2.Nudiviridae).
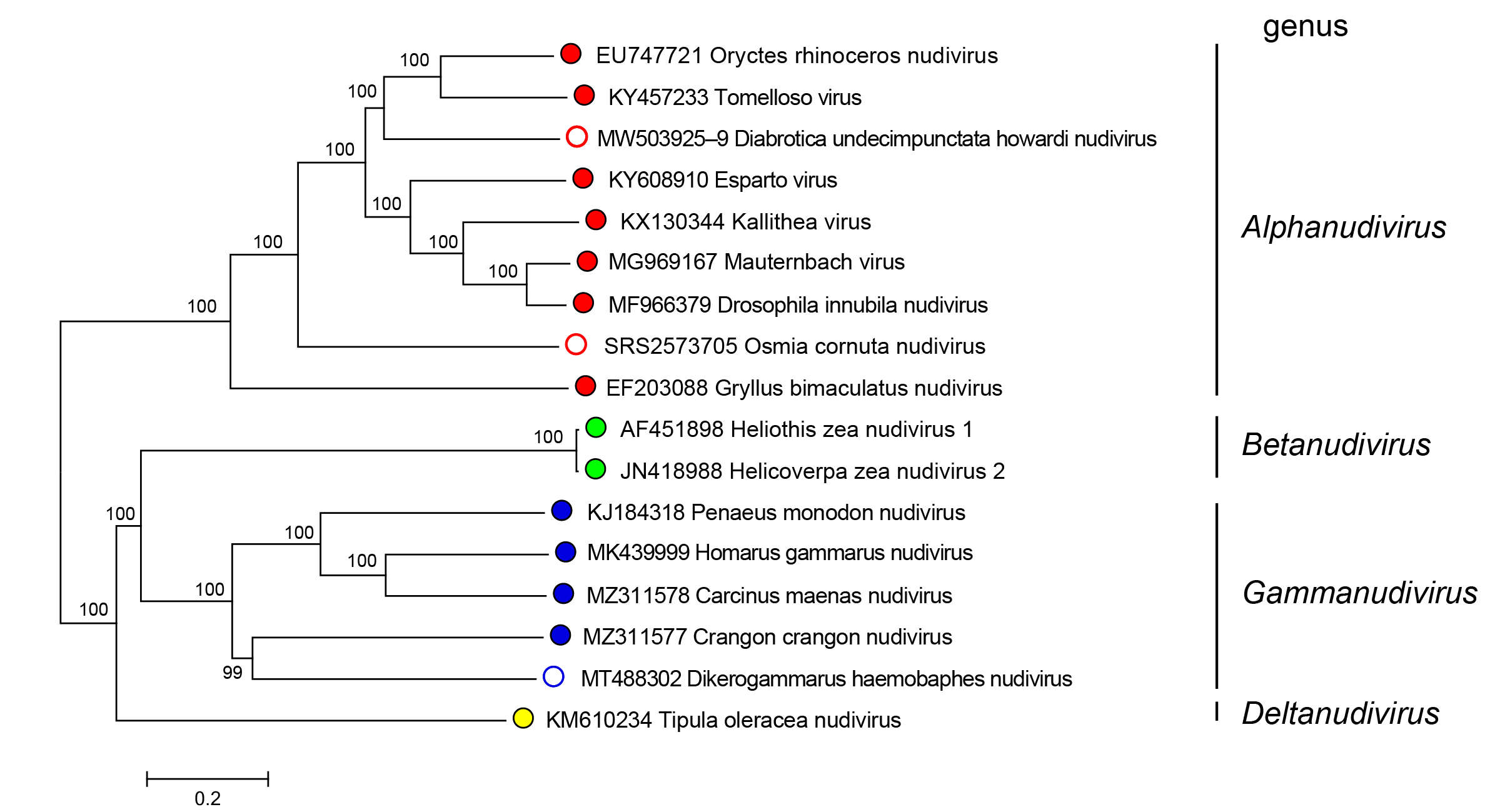 |
|
Figure 2.Nudiviridae. Phylogeny of members of the family Nudiviridae. The tree was constructed using 19 nudivirus core genes (38K, ac81, dnapol, helicase, helicase-2, integrase, lef-4, lef-8, lef-9, p47, p74/ pif-0, pif-1, pif-2, pif-3, pif-4, odv-e56/pif-5, vlf-1, vp39 and vp91/95) from 17 published nudiviruses (classified and no yet classified). Amino acid alignments were individually performed with default parameters using the MAFFT alignment plugin v7.0450 from Geneious Prime 2019.2.3 software. A consensus tree was then constructed at NGPhylogeny.fr website on concatenated and refined multiple alignments with parameters set as default parameters except for: Tree inference = PhyML, Evolutionary model = WAG, Equilibrium frequencies = Empirical, Tree topology search = Best of NNI and SPR, Statistical test for branch support = Bootstrap. Numbers on tree nodes indicate bootstrap supports (1000 replicates). Midpoint rooting. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Historically, nudiviruses have been classified within the family Baculoviridae as “non-occluded baculoviruses” but were removed from Baculoviridae upon the publication of the 6th Report in 1995 due to differences in virion morphology and the lack of occlusion bodies. Phylogenetic analysis based on the core genes shared with baculoviruses further demonstrated that nudiviruses comprise a monophyletic group distinguishable from baculoviruses (Wang et al., 2007). Therefore, these viruses were classified together into the family Nudiviridae, were grouped together into a family with two genera in 2013, expanded in 2021 to the current four genera. Nevertheless, nudiviruses share structural, genetic and biological characters with viruses of the family Baculoviridae. Nudiviruses share 21 core genes with baculoviruses (Wang et al., 2011). Nudiviruses are the closest evolutionary relatives of baculoviruses and there is evidence that these two families separated in the Paleozoic era (Thézé et al., 2011, Petersen et al., 2022). Nudiviruses are also related to viruses in the family Hytrosaviridae. These three virus families together form the order Lefavirales within the virus class Naldaviricetes that harbours all DNA viruses characterised by the presence of pif genes, encoding per os infectivity factors.
In addition, members of the genus Bracoviriform in the family Polydnaviriformidae derive from a mesozoic nudivirus that integrated into the genome of a parasitoid wasp (Herniou et al., 2013). In several other insect species, endogenised genomes of unclassified nudiviruses have been discovered, that appear to have integrated more recently. An example is the Nilaparvata lugens endogenous nudivirus in the brown planthopper that retained all twenty-eight nudivirus core genes displayed in Table 2.Nudiviridae (Cheng et al., 2014).

