Family: Hytrosaviridae
Henry M. Kariithi, Just M. Vlak, Johannes A. Jehle, Max Bergoin, Drion G. Boucias and Adly M. M. Abd-Alla
The citation for this ICTV Report chapter is the summary published as Kariithi et al., (2019):
ICTV Virus Taxonomy Profile: Hytrosaviridae, Journal of General Virology, 100, 1271–1272.
Corresponding author: Adly M. M. Abd-Alla (a.m.m.abd-Alla@iaea.org)
Edited by: Balázs Harrach and Sead Sabanadzovic
Posted: June 2019, updated May 2023
PDF: ICTV_Hytrosaviridae.pdf
Summary
Hytrosaviruses (also known as salivary gland hypertrophy viruses, SGHVs) are large, rod-shaped, enveloped, pathogenic viruses of dipterans with circular dsDNA genomes of 124–190 kbp (Table 1. Hytrosaviridae). Hytrosaviruses infect the hematophagous tsetse flies and the filth-feeding houseflies and are classified into two genera, Glossinavirus (with the single species Glossinavirus glopallidipedis) and Muscavirus (with the single species Muscavirus musdomesticae) respectively. Members of both genera, Glossina pallidipes salivary gland hypertrophy virus (infecting the tsetse fly) and Musca domestica salivary gland hypertrophy virus (infecting the housefly), have been fully sequenced. Members of the two genera have distinct genome sizes, host ranges and transmission modes. Hytrosaviruses primarily replicate in the salivary glands (SGs) of adult flies, thereby inducing SG hypertrophy syndrome (SGH). SGHVs, also replicate in other host tissues (e.g. trachea, milk glands, corpora allata/cardiaca) in which they do not induce hypertrophy. Muscavirus infections are strictly overt, while glossinavirus infections can be covert or overt. SGH-positive flies are almost always infertile. PCR-based estimates of the prevalence of hytrosaviruses in the field range from 1–88%. An unclassified hytrosavirus has been reported in the phytophagous narcissus bulb fly Merodon equestris.
Table 1. Hytrosaviridae. Characteristics of members of the family Hytrosaviridae
| Characteristic | Description |
|---|---|
| Example | Glossina pallidipes salivary gland hypertrophy virus (EF568108), species Glossinavirus glopallidipedis, genus Glossinavirus |
| Virion | Typically, enveloped particles of 50–80 × 500–1000 nm |
| Genome | Circular, dsDNA, 124–190 kbp, encoding 108–174 proteins |
| Replication | DNA synthesis and transcription within nuclear replication complexes; temporal expression of genes; nucleocapsids exit the nucleus via nuclear pore complex, then associate with the Golgi apparatus resulting in cytoplasmic envelopment and virion assembly |
| Translation | Presumably via cap-dependent, polyadenylated monocistronic mRNAs |
| Pathogenicity | Infection induces salivary gland hypertrophy (SGH) syndrome, reproductive dysfunctions and sterility in flies |
| Host range | Dipterans: tsetse flies spp. (Glossinavirus); housefly and stable fly (Muscavirus) |
| Taxonomy | Two genera (Glossinavirus and Muscavirus) each including a single species |
Virion
Morphology
Virions are 50–80 nm in diameter, non-occluded, non-icosahedral rod-shaped particles (500–1000 nm in length) and contain a bilayer lipid envelope (Abd-Alla et al., 2009, Garcia-Maruniak et al., 2009, Kariithi et al., 2013b, Orlov et al., 2018) (Figure 1A. Hytrosaviridae and 1D. Hytrosaviridae). Particles are bipolar with rounded and/or conical ends (Kariithi et al., 2013b, Orlov et al., 2018, Garcia-Maruniak et al., 2008) (Figure 1B. Hytrosaviridae and 1C. Hytrosaviridae). The dense internal capsid shows a helical organization and the envelope contains virus-encoded proteins with protruding, regularly-spaced and helically oriented glycoprotein spikes, presumably composed of protein dimers or braided, bead-like structures (Kariithi et al., 2013b, Orlov et al., 2018, Kariithi et al., 2010). Descriptions of members of the two individual hytrosavirus genera are detailed in the corresponding sections.
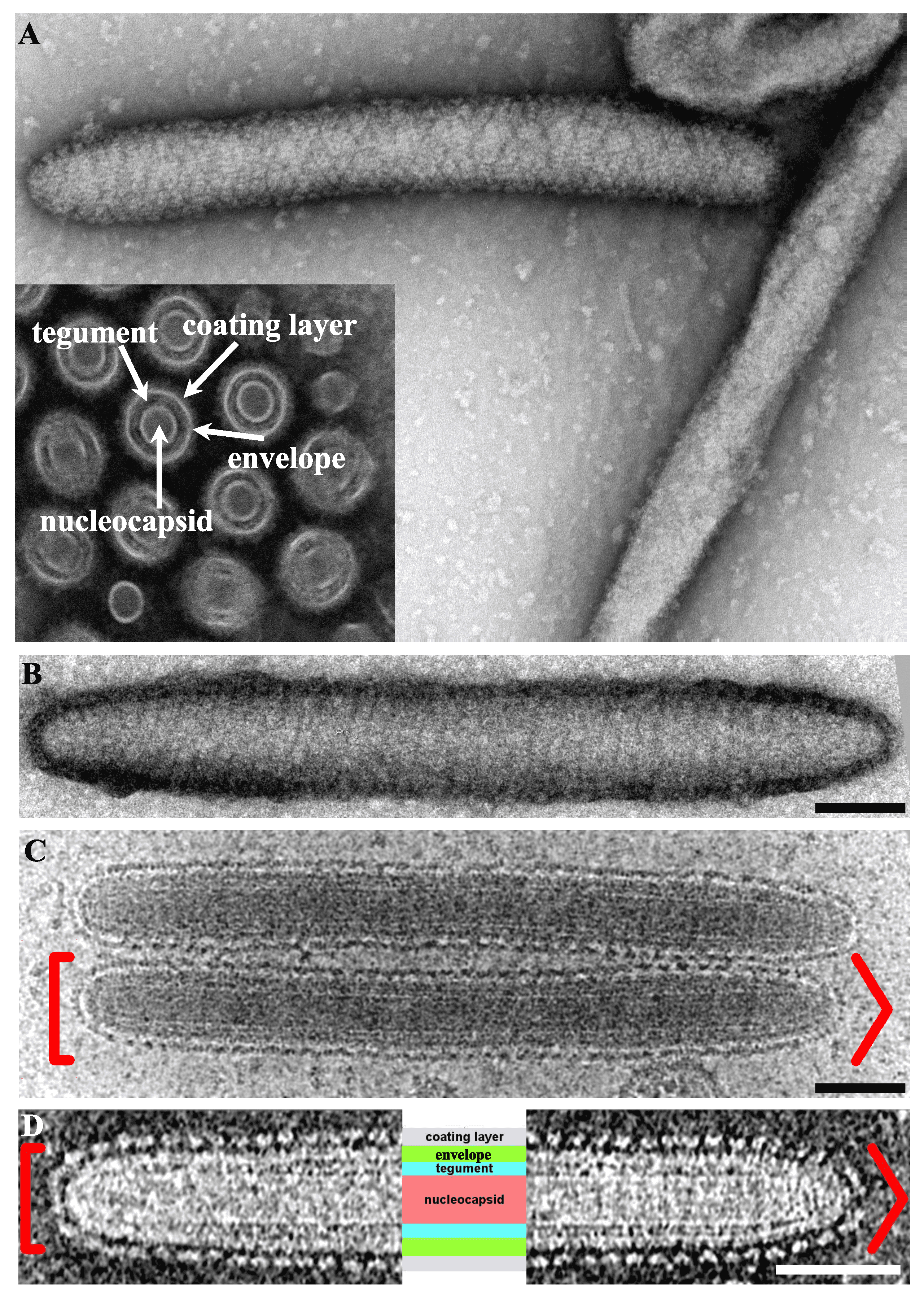 |
| Figure 1. Hytrosaviridae. Micrographs showing the structural features of Glossina pallidipes salivary gland hypertrophy virus – Uganda. (A) Transmission electron micrographs showing the main structural features of a virus particle. Inset in Panel A shows a cross-section indicating the nucleocapsid core, tegument, envelope and coating layer; (B) Negative-staining of a virion showing the helical organization of the coating protein. (C and D) Cryo-EM tomograms of the central section of virions exhibiting the same polarity with round (left [) and conical (right >) extremities. The four core parts of the virion including the spikes forming the coating layer, the envelope, the tegument and the nucleocapsid are labelled in light purple, green, turquoise and salmon, respectively. All scale bars in panels B–D are 100 nm. Micrographs in Panels A and B–D were adapted from (Kariithi et al., 2017a) and (Orlov et al., 2018), respectively. |
Physicochemical and physical properties
Virus particles have a density of 1.153 g cm-3 in Nycodenz gradients.
Nucleic acid
The hytrosavirus genome is a single copy of circular dsDNA of approximately 124 kbp (Muscavirus) or 190 kbp (Glossinavirus) (Garcia-Maruniak et al., 2009, Garcia-Maruniak et al., 2008, Abd-Alla et al., 2008, Abd-Alla et al., 2016). The majority of the hytrosavirus genes have 5′-terminal TATA-box promoter sequences, immediate early promoters with the CAGT promoter motif, the canonical baculovirus late (T/G/A)TAAG transcriptional initiation motifs and a 3′-terminal polyadenylation signal (Garcia-Maruniak et al., 2009, Abd-Alla et al., 2016).
Proteins
Hytrosavirus genomes contain 108 (Muscavirus) or 160–174 (Glossinavirus) putative genes (Garcia-Maruniak et al., 2008, Abd-Alla et al., 2008, Abd-Alla et al., 2016). Members of the genera Muscavirus and Glossinavirus encode at least 29 and 57 proteins, respectively, which are associated with the nucleocapsid, tegument and the envelope fractions of virus particles (Kariithi et al., 2013b, Garcia-Maruniak et al., 2008, Abd-Alla et al., 2016). Glossinavirus virions also contain 51 host-derived (cellular) proteins, 38 of which are associated with the viral tegument and the remaining 13 are potentially incorporated into virions (Kariithi et al., 2013b).
Lipids
Hytrosaviruses acquire their lipid envelopes through cytoplasmic assembly by budding through endoplasmic/Golgi membranes (Kariithi et al., 2013b, Orlov et al., 2018, Boucias et al., 2013a, Lietze et al., 2011c).
Carbohydrates
Unknown.
Genome organization and replication
All hytrosaviruses have a similar organization of their circular dsDNA genome. The genome of members of the genus Glossinavirus (190 kbp) is about 65 kbp longer than that of members of the genus Muscavirus (124 kbp), contains 52–66 more putative non-overlapping genes and has a significantly lower G+C content (28% vs. 44%) (Garcia-Maruniak et al., 2009, Garcia-Maruniak et al., 2008, Abd-Alla et al., 2008, Abd-Alla et al., 2016). The conserved regions in the genome of the two known glossinavirus strains are 98.1% identical at the nucleotide level, compared to a lower collinearity (25–73%) between the corresponding regions in the muscavirus genome (Figure 2. Hytrosaviridae) (Abd-Alla et al., 2009, Garcia-Maruniak et al., 2009, Abd-Alla et al., 2016). Genes are distributed equally over both DNA strands and occur in unidirectional clusters (Figure 3. Hytrosaviridae). Approximately 2.8% and 1.7% of the glossinavirus and muscavirus genomes, respectively, contain homologous repeat sequences (hrs). The glossinavirus genome contains a total of 14 direct repeats sequence (drs; composed of 51–246 bp units) compared to 18 drs in the muscavirus genome (composed of 9–149 bp units). In addition, one inverted repeat sequence occurs in the glossinavirus genome (but not in the muscavirus genome). The drs are clustered in certain regions on the genomes, but they are less clustered in muscaviruses than in glossinaviruses (Garcia-Maruniak et al., 2009, Garcia-Maruniak et al., 2008, Abd-Alla et al., 2008, Abd-Alla et al., 2016). In contrast to other large invertebrate dsDNA viruses, approximately 50% of the drs are localized within specific ORFs and are thought to be origins of viral DNA replication and/or act as transcriptional enhancers.
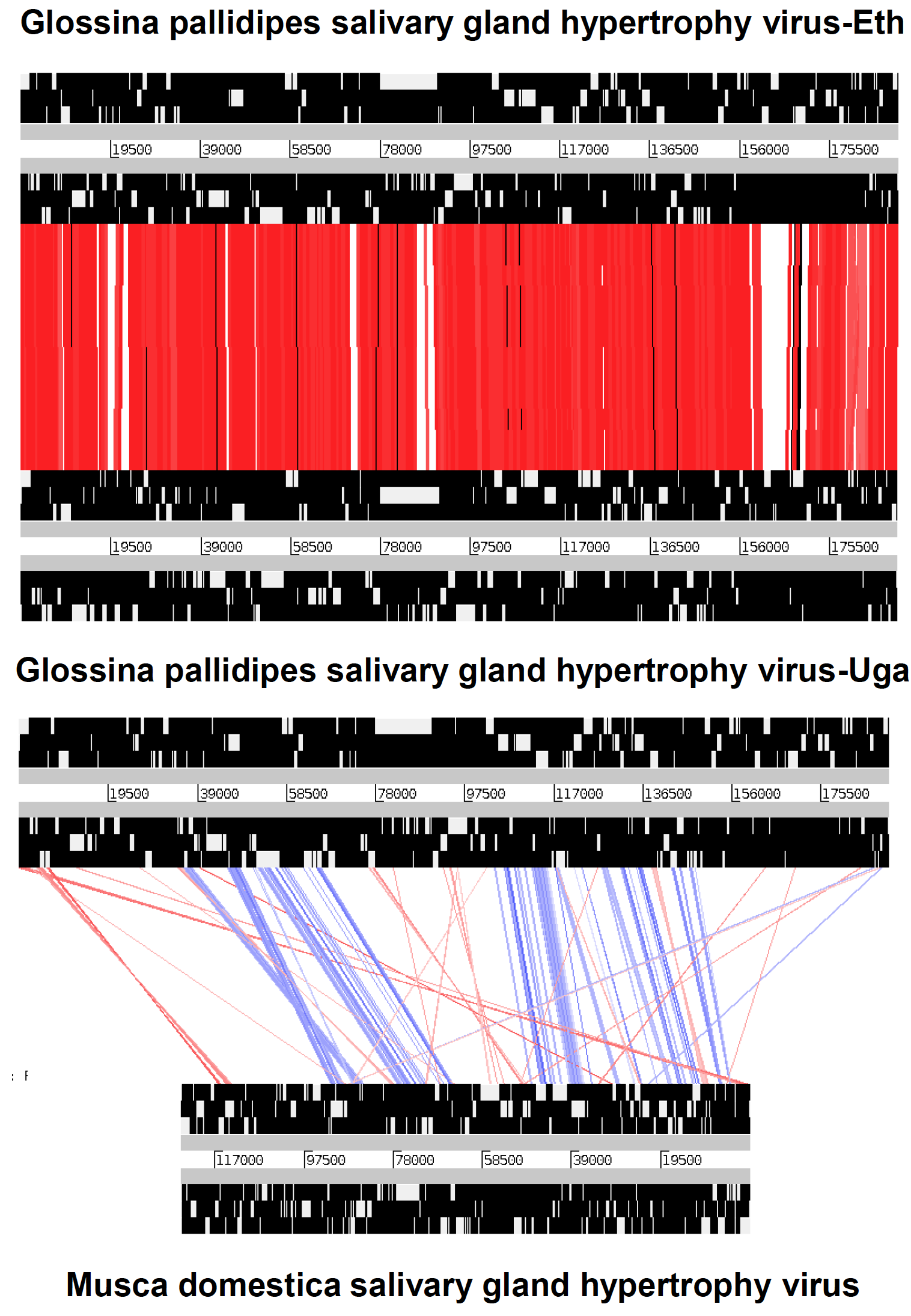 |
| Figure 2. Hytrosaviridae. Collinearity of the conserved regions of Ugandan and Ethiopian glossinavirus isolates (GpSGHV-Eth and GpSGHV-Uga, A) compared with muscavirus (B): The red lines in A and B) indicate identity levels of regions (not ORF) between the viruses, whilst the blue lines (in B) indicate inversions. The white bands represented in the thick black lines do not necessarily indicate the open reading frames (ORFs), but rather the conserved genomic regions. The figure was adapted from (Abd-Alla et al., 2016). |
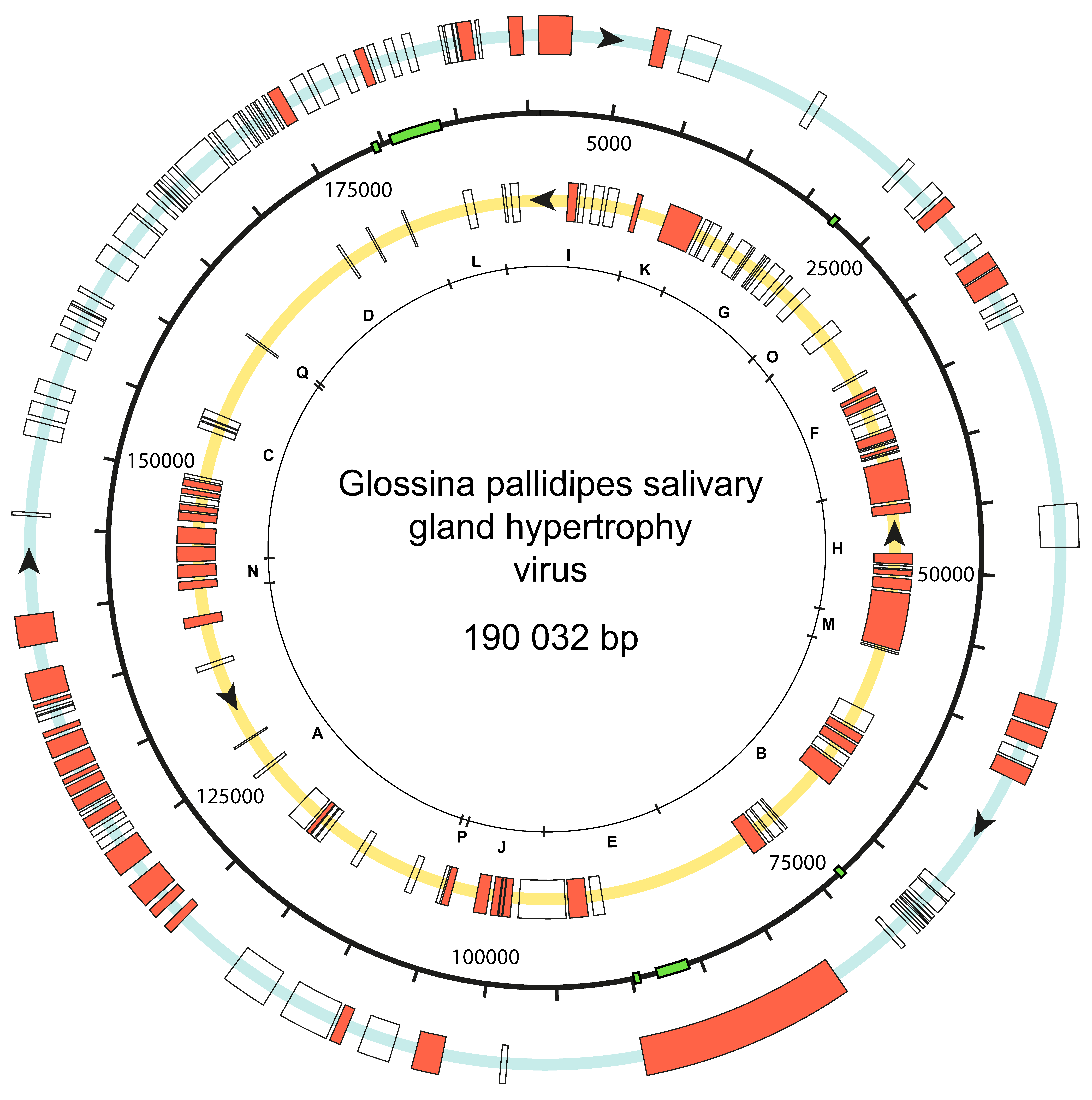 |
| Figure 3. .Hytrosaviridae. Circular representation of the genome of Glossina pallidipes salivary gland hypertrophy virus (Ethiopia strain): Putative open reading frames are indicated by boxes on the blue (clockwise transcription) or yellow (anti-clockwise transcription) circles, with those encoding virion proteins indicated by salmon colour. Alphabetical numbers on the innermost ring represent BglII restriction fragments, while green boxes on the genome circle indicate the position of the direct repeat sequences. Information derived from (Abd-Alla et al., 2016). |
Hytrosaviruses primarily replicate in viral replication complexes in the nuclei of the host’s SGs cells, but also in non-SG tissues (e.g. milk glands, corpora allata/cardiaca) (Garcia-Maruniak et al., 2009, Kariithi et al., 2017b). Orally ingested hytrosaviruses exploit the tracheal system as a conduit to breach basal laminal barrier in the midgut to access other tissues including the SGs (Lietze et al., 2011c). Infection is thought to initiate via attachment to unknown host receptors followed by internalization (Kariithi et al., 2011). The fusion of viral and host vesicle membranes eventually results in the release of the capsids into the cytoplasm, which are then trafficked to the cell nucleus where the genome is released for gene transcription and genome replication, packaging and assembly of progeny nucleocapsids in the virogenic stroma (Kariithi et al., 2013b, Lietze et al., 2011c, Kariithi et al., 2011). Hytrosavirus replication is thought to involve the sequential expression of immediate early (transcription factors), early (DNA replication genes), and late (viral replication proteins) genes (Kariithi et al., 2013b). Nucleocapsids exit the nucleus via the nuclear pore complex. Virion assembly, including the acquisition of the glycoprotein-containing envelopes, occurs in the cytoplasm. Mature particles egress by budding through (Muscavirus) or lysis of the luminal membranes of infected cells (Glossinavirus) (Kariithi et al., 2013b, Boucias et al., 2013a, Lietze et al., 2011c), into the lumen of the salivary gland.
Biology
Known hytrosaviruses are restricted to three dipteran insects; the hematophagous tsetse flies (Glossina spp.), the filth-feeding housefly, Musca domestica, and the phytophagous syrphid fly Merodon equestris (Abd-Alla et al., 2009). The distinct biological characteristics of members of the genera Glossinavirus and Muscavirus are described in the corresponding genus pages.
Pathogenicity
The salient diagnostic feature of hytrosavirus infection is the induction of salivary gland hypertrophy (SGH) syndrome in the SGs of infected flies (Figure 4A. Hytrosaviridae and 4C. Hytrosaviridae) (Abd-Alla et al., 2009). Muscavirus-infected SGs are hypertrophic (i.e. contain uniformly infected cells with enlarged cytoplasmic and nuclear compartments) (Figure 4B. Hytrosaviridae), while glossinavirus-infected SGs are hyperplastic (i.e. contain multi-layered dividing cells) (Figure 4D. Hytrosaviridae). Hytrosavirus infection causes reproduction dysfunctions and sterility (Kariithi et al., 2017b, Abd-Alla et al., 2010). The pathogenetic characteristics that distinguish members of the two Hytrosaviridae genera are described in the corresponding genus pages.
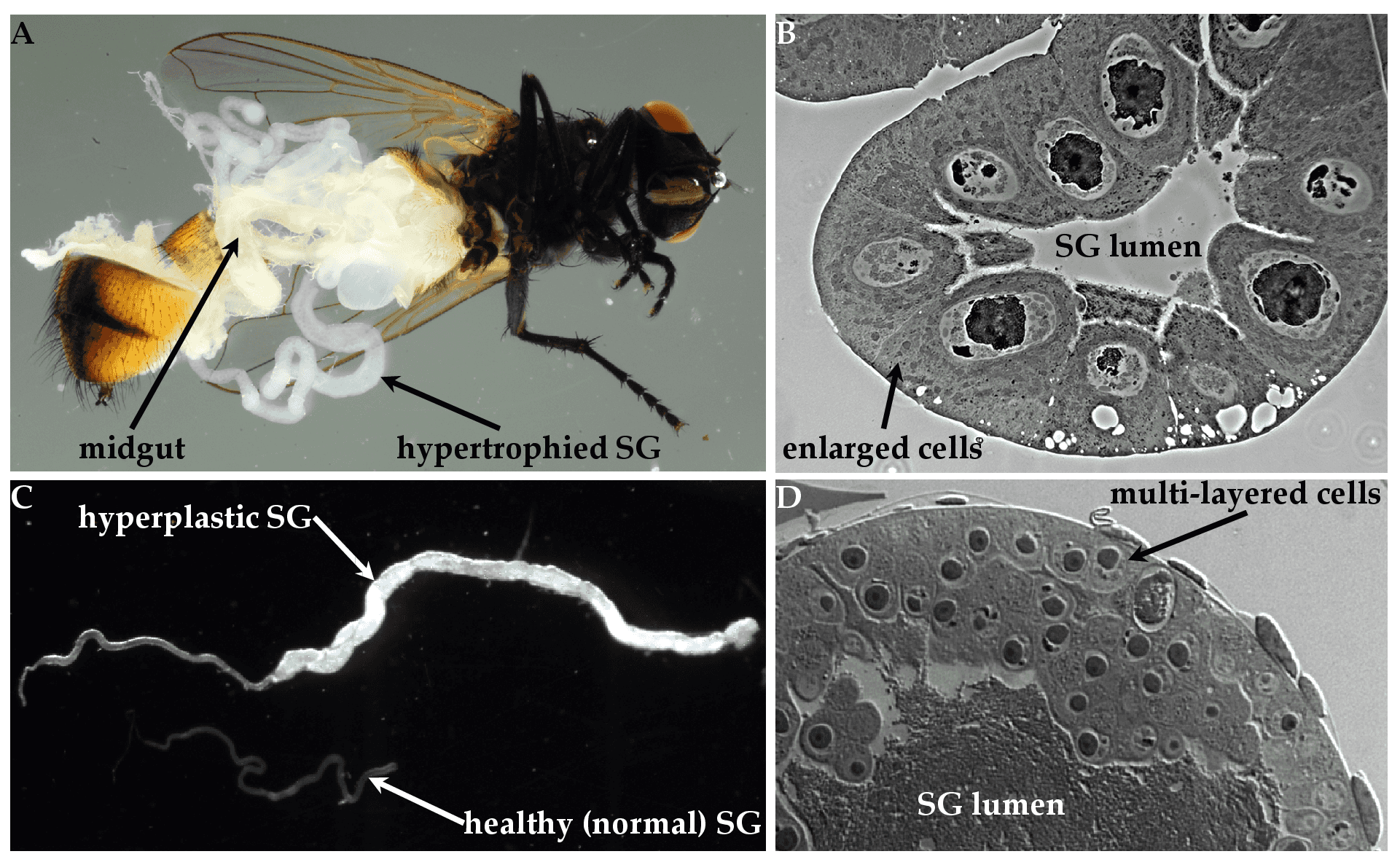 |
| Figure 4. Hytrosaviridae. Micrographs showing the pathogenicity of salivary gland hypertrophy viruses: (A) Light micrographs of hypertrophied salivary glands (SGs) dissected from Musca domestica infected by muscavirus. (B) Muscavirus-induced cellular hypertrophy in housefly SGs. (C) Hyperplastic SGs of tsetse. (D) Glossinavirus-induced hyperplasia in tsetse SGs. Figure source: (Kariithi et al., 2017a). |
Antigenicity
Various proteins including the major envelope protein p74 of glossinaviruses have been identified as immunogenic in rabbit (Kariithi et al., 2010).
Derivation of names
Hytrosaviridae: from the Greek Hypertrophia meaning excess nourishment and sialoadenitis meaning salivary gland inflammation (Abd-Alla et al., 2009).
Relationships within the family
Phylogenetically, the 190 kbp genome of glossinavirus has limited, but significant gene homology to the muscavirus 124 kb genome (Garcia-Maruniak et al., 2008, Abd-Alla et al., 2008, Abd-Alla et al., 2016), hence the placement of the hytrosaviruses into two separate genera within the family Hytrosaviridae. Based on their virus particle dimensions, members of the genus Muscavirus are more similar than those of the genus Glossinavirus to an unclassified virus that has been reported to infect the narcissus bulb fly Merodon equestris. This virus is a putative member of the Hytrosaviridae on the basis of similarity of virus particles and SGH symptomatology (Amargier et al., 1979).
Relationships with other taxa
Hytrosaviruses are structurally similar to baculoviruses, nudiviruses and nimaviruses, but differ functionally in the absence of occlusion bodies, their host ranges and predominant modes of transmission, as well as lower lethality (i.e. hytrosaviruses rarely kill their insect host) (Kariithi et al., 2018). Based on phylogenetic analysis of the DNA polymerase (dnapol) gene, which is present in all large dsDNA viruses, hytrosaviruses genomes cluster more closely with linear dsDNA genomes than with the large circular invertebrate dsDNA viruses (Figure 5. Hytrosaviridae) (Garcia-Maruniak et al., 2008, Abd-Alla et al., 2008). Among the structural and genomic features shared by hytrosaviruses and some other large dsDNA viruses are the possession of enveloped, rod-shaped virions, circular dsDNA genome, the presence of multiple interspersed direct homologous repeats and replication in the nucleus. Members of the Hytrosaviridae share 12 of the 38 core genes that have been described in baculoviruses (Javed et al., 2017, Wennmann et al., 2018). The most significant similarity between hytrosaviruses and other large invertebrate dsDNA viruses is the presence of the per os infectivity factor genes (p74/pif-0, pif-1, pif-2 and pif-3) in members of both Muscavirus and Glossinavirus (Figure 6. Hytrosaviridae). These genes are conserved in baculoviruses, nudiviruses, nimaviruses and some bracoviruses (Jehle et al., 2013), and suggest a common mechanism of host entry. Hytrosaviruses possess genes encoding several subunits of the viral DNA-directed RNA polymerase (DdRP) complex, including four late expression factor genes (lef-4, lef-5, lef-8 and lef-9) found in baculoviruses and nudiviruses (Abd-Alla et al., 2016, Jehle et al., 2013). However, lef-3 was not found in hytrosaviruses.
 |
| Figure 5. Hytrosaviridae. Phylogenetic tree of hytrosaviruses based on predicted DNA polymerase amino acid sequences. The phylogenetic tree of DNA polymerase and its homologs is based on 603 conserved aa sites in 30 viruses from various families. The evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix in MEGA7 (Kumar et al., 2016). A discrete Gamma distribution was used to model evolutionary rate differences among sites (2 categories (+G, parameter = 1.5222)). All positions containing gaps and missing data were eliminated. Numbers on the nodes indicate bootstrap values (500 replicates). |
|
| Figure 6. Hytrosaviridae. Phylogenetic tree of concatenated predicted amino acid sequences of per os infectivity factor proteins (P74, PIF-1, PIF-2, PIF-3 and ODV-E66) of hytrosaviruses and their homologues in baculoviruses and nudiviruses. The evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix model in MEGA7 (Kumar et al., 2016). A discrete Gamma distribution was used to model evolutionary rate differences among sites with categories and a G parameter of 1.7509 (A: Top) or 0.9778 (B: Bottom). All positions containing gaps and missing data were eliminated. Numbers on the nodes indicate bootstrap values (500 replicates). The figure was adapted from (Garcia-Maruniak et al., 2009). |



