Family: Caulimoviridae
Pierre-Yves Teycheney, Andrew D.W. Geering, Idranil Dasgupta, Roger Hull, Jan F. Kreuze, Ben Lockhart, Emmanuelle Muller, Neil Olszewski, Hanu Pappu, Mikhail M. Pooggin, Katja R. Richert-Pöggeler, James E.Schoelz, Susan Seal, Livia Stavolone and Marie Umber
The citation for this ICTV Report chapter is the summary published as Teycheney et al., (2020):
ICTV Virus Taxonomy Profile: Caulimoviridae, Journal of General Virology 101:1025–1026
Corresponding author: Pierre-Yves Teycheney (teycheney@cirad.fr)
Edited by: F. Murilo Zerbini and Sead Sabanadzovic
Posted: August 2020
PDF: ICTV_Caulimoviridae.pdf
Summary
Caulimoviridae is a family of non-enveloped reverse-transcribing plant viruses with non-covalently closed circular dsDNA genomes of 7.1–9.8 kbp that are encapsidated by viral coat proteins into isometric virions (members of the genera Caulimovirus, Cavemovirus, Petuvirus, Rosadnavirus, Solendovirus and Soymovirus) or bacilliform-shaped virions (members of the genera Badnavirus and Tungrovirus) (Table 1. Caulimoviridae). No data is currently available on the virion morphology for viruses in the genera Dioscovirus and Vaccinivirus. Members of the Caulimoviridae infect a wide range of monocot and dicot plants, although members of a single species have generally narrow host ranges. Some viruses of this family cause economically important diseases of tropical and subtropical crops. Insect vectors have been identified for badnaviruses, caulimoviruses and tungroviruses. Endogenous viral elements (EVEs; viral DNA integrated into the host nuclear genome) are known for badnaviruses, caulimoviruses, cavemoviruses, petuviruses and solendoviruses. Some caulimovirid EVEs in Musa balbisiana, Petunia hybrida and Nicotiana edwardsonii are replication competent and can lead to spontaneous infections by episomal forms of the viral genome through activation by biotic or abiotic stresses. However, the majority of caulimovirid EVEs, which are widespread in the genomes of vascular plants, are considered to be replication defective.
Table 1. Caulimoviridae. Characteristics of members of the family Caulimoviridae
|
Characteristic |
Description |
|
Typical member |
cauliflower mosaic virus-Cabb-S (V00141), species Cauliflower mosaic virus, genus Caulimovirus |
|
Virion |
Virions are either isometric or bacilliform with no envelope |
|
Genome |
Non-covalently closed circular dsDNA of 7.1–9.8 kbp with discontinuities in both strands |
|
Replication |
Nuclear transcription by host DNA-directed RNA polymerase II, generates pregenomic (pg)RNA and, in members of some genera, subgenomic (sg)RNA. Replication is cytoplasmic via reverse transcription of pgRNA by viral reverse transcriptase |
|
Translation |
From capped and polyadenylated pgRNA and, in members of some genera, sgRNA and spliced version(s) of pgRNA |
|
Host range |
Plants (monocots and dicots) |
|
Taxonomy |
Realm Riboviria¸ kingdom Pararnavirae, phylum Artverviricota, class Revtraviricetes, order Ortervirales; ten genera including 85 species |
Virion
Morphology
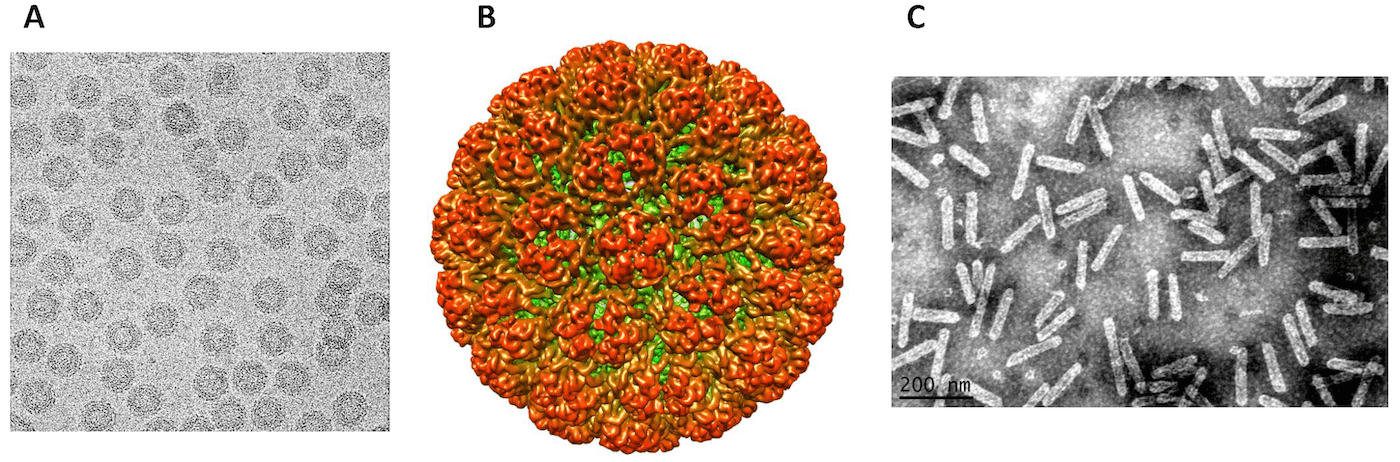
Virions are either isometric or bacilliform depending on the genus (Figure 1. . Caulimoviridae). There is no envelope.
Physicochemical and physical properties
Virions have buoyant densities in CsCl of 1.37 g cm−3 (members with isometric virions) or in Cs2SO4 of 1.31 g cm−3 (members with bacilliform virions). S20,w is in the range of 200 to 220 S. Virions are very stable between pH 4 and 9 and in high salt concentrations.
Nucleic acid
Virions contain a single molecule of non-covalently closed circular dsDNA of 7.1–9.8 kbp. Each strand of the genome has discontinuities at specific sites (positions of the primers for reverse transcriptase): the negative-sense strand has one discontinuity (Met-tRNA primer binding site) and the positive-sense strand has between one and three discontinuities (purine-rich sequences resistant to RNaseH).
Proteins
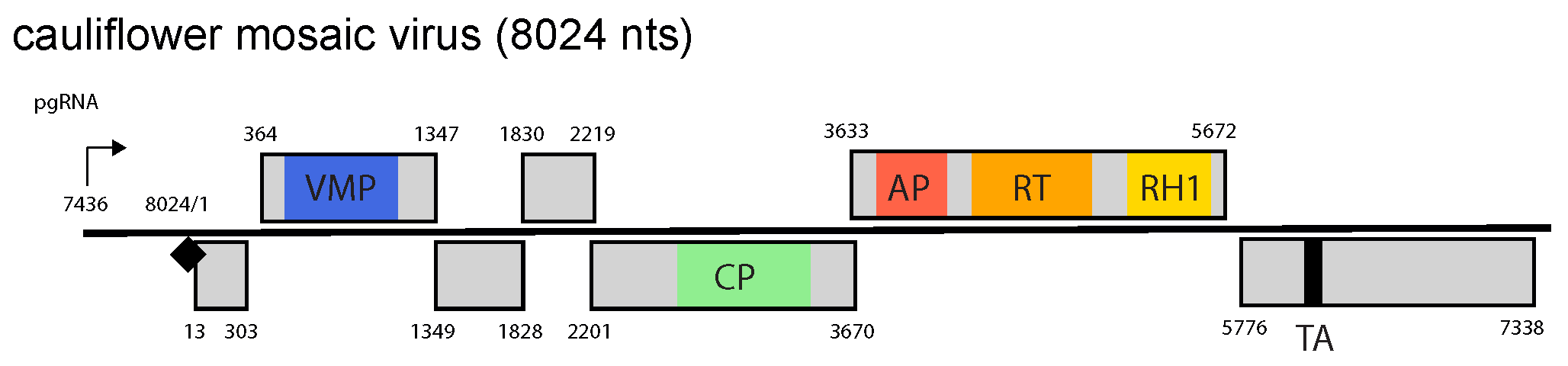
Genomes contain between one and eight ORFs, depending on the genus. The virus-encoded proteins common to all viruses, regardless of the genus they belong to, are a movement protein (MP), a capsid protein (CP), a multipurpose virion-associated protein (VAP), an aspartic protease (AP) and a reverse transcriptase (RT) with tethered RNAse H1 enzyme (Figure 2. Caulimoviridae).
 |
|
Figure 2. Caulimoviridae. Genome organization of cauliflower mosaic virus. The linearized map begins at the pgRNA transcription start site (black arrow, mapped or predicted ca. 32 nts downstream of TATA box; see (Pooggin et al., 1999) and references therein). The numbering begins from the first nucleotide of the Met-tRNA primer binding site (black diamond). Light grey boxes mark open reading frames (ORFs). Conserved protein domains as listed in the Pfam database (http://xfam.org) are colored: blue is the viral movement protein (VMP) (PF01107), red is the retropepsin (pepsin-like aspartic protease) (AP) (CD00303), orange is the reverse transcriptase (RT) (CD01647) and yellow is the RNase H1 (RH1) (CD06222). The conserved C-terminus of the coat protein (CP) is marked green. The conserved translation transactivator (TA) domain is shown in black. |
Lipids
None.
Carbohydrates
The capsid protein of cauliflower mosaic virus (CaMV) is glycosylated.
Genome organization and replication
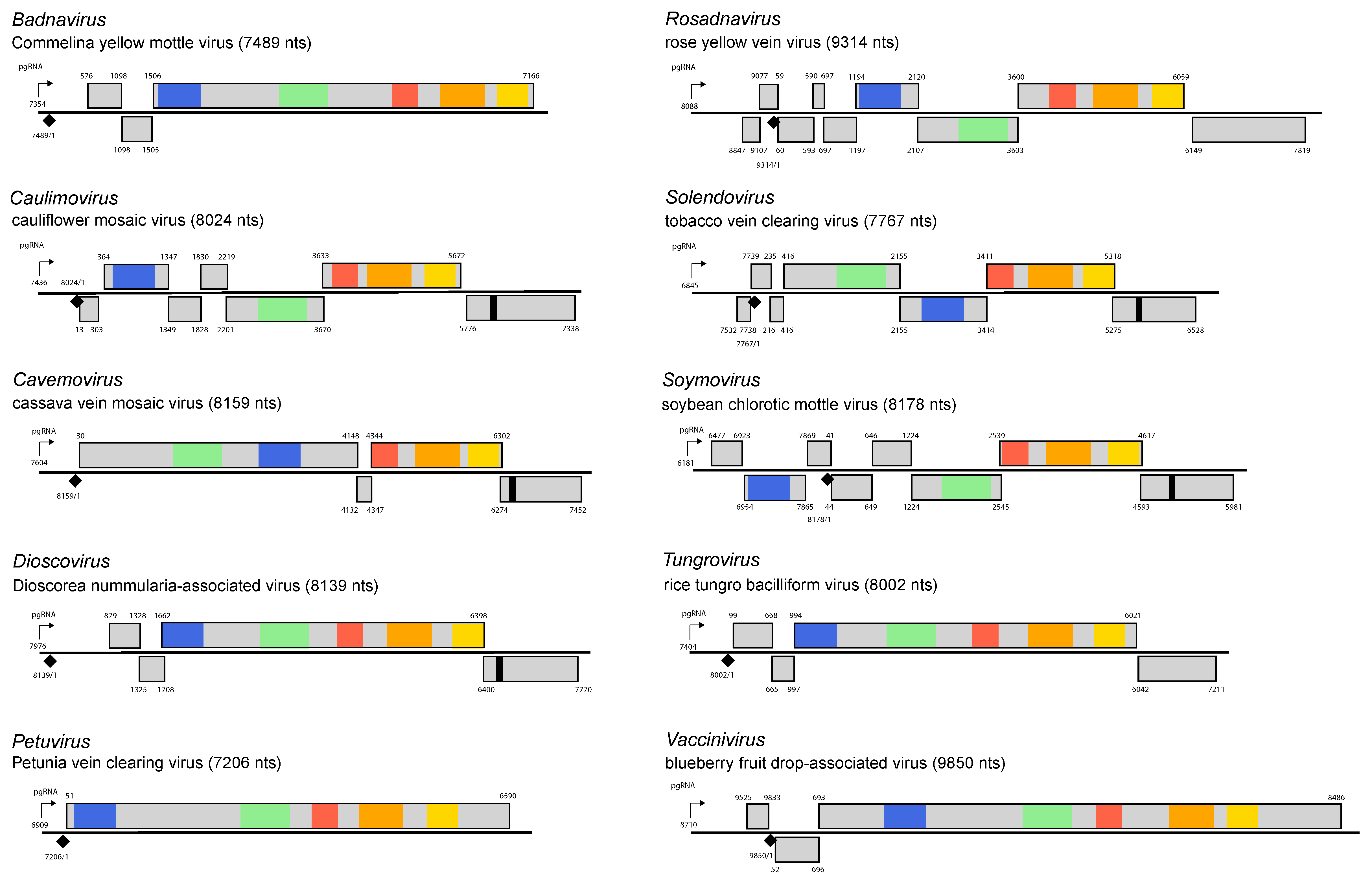
By convention, numbering of the genome starts at the 5′-end of the negative-sense strand primer-binding site. One strand of DNA contains the coding sequence (positive-sense strand). The genome organization is genus-specific (Figure 3. Caulimoviridae) and distinguishes members of different genera from each other.
 |
|
Figure 3. Caulimoviridae. Genome organization of type members of the genera Badnavirus, Caulimovirus, Cavemovirus, Dioscovirus, Petuvirus, Rosadnavirus, Soymovirus, Solendovirus, Tungrovirus and Vaccinivirus. The linearized map begins at the pgRNA transcription start site (black arrow, mapped or predicted ca. 32 nts downstream of TATA box; see (Pooggin et al., 1999) and references therein). The numbering begins from the first nucleotide of the Met-tRNA primer binding site (black diamond). Light grey boxes mark open reading frames (ORFs). Conserved protein domains as listed in the Pfam database (http://xfam.org) are colored: blue is the viral movement protein (PF01107), red is the retropepsin (pepsin-like aspartic protease) (CD00303), orange is the reverse transcriptase (CD01647) and yellow is the RNase H1 (CD06222). The conserved C-terminus of the coat protein is marked green. The conserved translation transactivator (TA) domain is shown in black. Note that in Commelina yellow mottle virus, an authentic start codon of ORF1 is the second in-frame AUG at position 576 (Cheng et al., 1996), consistent with position of the ribosome shunt landing site (Pooggin et al., 1999). In rice tungro bacilliform virus, ORF1 begins with a non-AUG start codon AUU at position 99 (Fütterer et al., 1996). For the type members of Dioscovirus and Vaccinivirus, the pgRNA leader region was defined based on the presence of a shunt configuration with the 5′-proximal short ORF in front of a large hairpin-like secondary structure (M. Pooggin, unpublished sequence analysis), which is a conserved feature in the family Caulimoviridae (Pooggin et al., 1999, Pooggin and Ryabova 2018). With the exception of petuviruses containing a single large ORF, three or more consecutive ORFs following the pgRNA leader region are likely translated via leaky scanning or TA-mediated reinitiation as has been established for members of the genera Tungrovirus and Caulimovirus, respectively, and predicted for members of other genera (Pooggin and Ryabova 2018) (M. Pooggin, unpublished sequence analysis) for members of the genera Dioscovirus and Vaccinivirus. |
Following entry into the cell, the virion is targeted to the nucleus by a nuclear localization signal (NLS) that is located in the N-terminus of the capsid protein and exposed on the surface of the virion (Karsies et al., 2002). It is then thought that the virion docks at a nuclear pore, virion disassembly occurs and the DNA is targeted to the nucleus utilizing the importin α pathway or perhaps other pathways in the case of rice tungro bacilliform virus (RTBV) (Guerra-Peraza et al., 2005). The discontinuities in the genome are then sealed to give supercoiled DNA, which associates with histone proteins to form mini-chromosomes in the nucleus. These are transcribed asymmetrically by the host DNA-directed RNA polymerase II (Pol II) to give a greater-than-genome length transcript (35S or 34S RNA) that has a terminal redundancy of about 35 to 270 nt, dependent upon the species. This transcript serves as a template (the pregenomic RNA) for reverse transcription which produces the minus-strand DNA , and as a polycistronic mRNA for expression of at least some of the ORFs (Pooggin and Ryabova 2018, Hohn and Richert-Pöggeler 2006).
The 5′-leader sequence of the pregenomic RNA of nearly all members of the Caulimoviridae is long and folds into a large and stable hairpin structure. Initiation of translation of the first large ORF occurs by a ribosome shunt mechanism, so bypassing this structure (Pooggin and Ryabova 2018). Viruses in the genus Caulimovirus produce a subgenomic 19S RNA, a monocistronic mRNA for ORF6 coding for a multifunctional transactivator/viroplasmin (TA) protein involved in transactivation of polycistronic translation, replication and suppression of antiviral defenses (Pooggin and Ryabova 2018). Viruses in the genera Cavemovirus, Rosadnavisus, Dioscovirus and Solednovirus encoding a TA homolog at the same distal position presumably express TA from a subgenomic RNA and share with caulimoviruses a reinitiation mechanism of polycistronic translation (Pooggin and Ryabova 2018). In the tungrovirus RTBV, the first three ORFs are translated from pgRNA via ribosome shunting and leaky scanning, while the spliced pregenomic RNA serves as a monocistronic mRNA for ORF4 coding for a defense suppressor protein (Rajeswaran et al., 2014). In the caulimovirus CaMV, one of the alternatively spliced versions of the pregenomic RNA serves as a polycistronic mRNA for ORF3 and further downstream ORFs (Pooggin and Ryabova 2018). No sgRNAs or spliced pgRNAs have been reported (or predicted) for viruses in the genera Badnavirus, Petuvirus and Vaccinivirus. Badnaviruses presumably translate three consecutive ORFs from pgRNA via ribosome shunting and leaky scanning similar to RTBV (Pooggin and Ryabova 2018), except for two of its members, Jujube mosaic-associated virus and sweet potato pakakuy virus, for which ORF3 is split in two. The petuvirus Petunia vein clearing virus (PVCV) and the vaccinivirus blueberry fruit drop-associated virus (BFDaV) have a single large ORF encoding a polyprotein precursor of all viral proteins (Figure 3. Caulimoviridae).
The replication cycle, in contrast to that of retroviruses, is episomal and does not involve an integration phase (Krupovic et al., 2018). Negative-sense strand DNA synthesis is primed by host cytosolic tRNAmet, and synthesis of both strands is performed by the viral reverse transcriptase (RT) and RNase H1. The site-specific discontinuities are at the priming sites for both negative-sense and positive-sense strand DNA synthesis and are made by the oncoming strand displacing the existing strand for a short distance and not ligating to form a closed circle (Hohn and Richert-Pöggeler 2006, Hohn and Rothnie 2013).
Biology
Most viruses have a narrow host range; those in the genera Caulimovirus, Cavemovirus, Petuvirus, Rosadnavirus, Solendovirus, Soymovirus and Vaccinivirus are restricted to dicotyledonous plants; tungroviruses and dioscoviruses infect monocotyledonous plants and badnaviruses infect either mono- or dicots. Many members of the family are spread by vegetative propagation.
The geographic range of many caulimovirids is wide; members of most species in the genera Badnavirus, Tungrovirus and Dioscovirus are primarily tropical or subtropical with some of them being widespread in temperate and sub-Antarctic climates, whereas members of most species in the genera Caulimovirus, Cavemovirus, Petuvirus, Rosadnavirus, Solendovirus, Soymovirus and Vaccinivirus are found in temperate regions.
The symptoms caused by these viruses are variable and dependent on the virus, host and climatic conditions. Mosaic or vein clearing symptoms predominate amongst members of the genera Caulimovirus, Cavemovirus, Petuvirus, Rosadnavirus, Solendovirus and Soymovirus whereas interveinal chlorotic mottling, stunting, yellow-orange foliar discoloration and streaking are the most frequent symptoms associated with infections by viruses in the genera Tungrovirus and Badnavirus.
Most viruses in the family infect most cell types of their hosts although some members of the genera Tungrovirus and Badnavirus are restricted to the phloem sieve tubes and xylem parenchyma. Virions occur in the cytoplasm and members of species in the genera Petuvirus, Caulimovirus, Soymovirus and Cavemovirus are associated with virus-encoded proteinaceous inclusion bodies. The movement protein (MP) of cauliflower mosaic virus uses the endocytic pathway to target the plasma membrane and forms tubules inside plasmodesmata, through which virions move from cell-to-cell. MP also facilitates long distance movement of the virus (Carluccio et al., 2014, Schoelz et al., 2016).
Members of the genera Badnavirus, Caulimovirus, Cavemovirus, Petuvirus and Solendovirus may have both endogenous (viral DNA integrated in the host nuclear genome) and exogenous episomal forms (Teycheney and Geering 2011). Endogenous viral elements (EVEs) resembling extant caulimovirids are widespread in the genomes of vascular plants, suggesting the past existence of additional viruses in the family, which now exist only as fossil sequences in their host’s genome (Geering et al., 2014, Diop et al., 2018). Integration is not an obligatory step in the replication cycles of these viruses but rather the DNA became captured in the host nuclear genome by non-homologous end-joining (also known as illegitimate recombination). Replication-competent endogenous forms of caulimovirids occur in the plant species Musa balbisiana, Petunia hybrida and Nicotiana edwardsonii (Teycheney and Geering 2011). Replication-defective endogenous caulimovirid elements are widespread in dicotyledonous and monocotyledonous plants (Geering et al., 2014, Diop et al., 2018).
Antigenicity
Virions of caulimovirids are moderate to efficient immunogens. There is pronounced antigenic variability between viruses belonging to different species in the genus Badnavirus (Vo et al., 2016). There are some serological cross-reactions between members of different genera.
Derivation of names
Badnavirus: from bacilliform DNA viruses.
Caulimovirus: from cauliflower mosaic virus.
Cavemovirus: from cassava vein mottle virus.
Dioscovirus: from Dioscorea, the genus containing D. nummularia, host to the type member.
Petuvirus: from petunia vein clearing virus.
Rosadnavirus: from Rosa DNA virus, where Rosa is the generic epithet of rose and DNA refers to the composition of the virus genome.
Solendovirus: from Solanaceae endogenous virus.
Soymovirus: from soybean chlorotic mottle virus.
Tungrovirus: from rice tungro bacilliform virus, tungro meaning “degenerated growth” in a Filipino dialect.
Vaccinivirus : from Vaccinium, the genus of the cultivated blueberry, host to the type member.
Genus demarcation criteria
Genera within the family Caulimoviridae are distinguished by:
- Virion morphology – members of the genera Badnavirus and Tungovirus have bacilliform virions whereas members of other genera have isometric virions.
- Genome organization: one ORF for members of genera Petuvirus and Vaccinivirus; 3 or 4 for members of the genera Badnavirus, Dioscovirus and Tungovirus, and 4–8 for members of other genera.
- Transmission mode and vector species.
- Host types – monocots for members of the genus Tungrovirus, dicots for other genera except for Badnavirus whose members infect both mono and dicots.
Relationships within the family
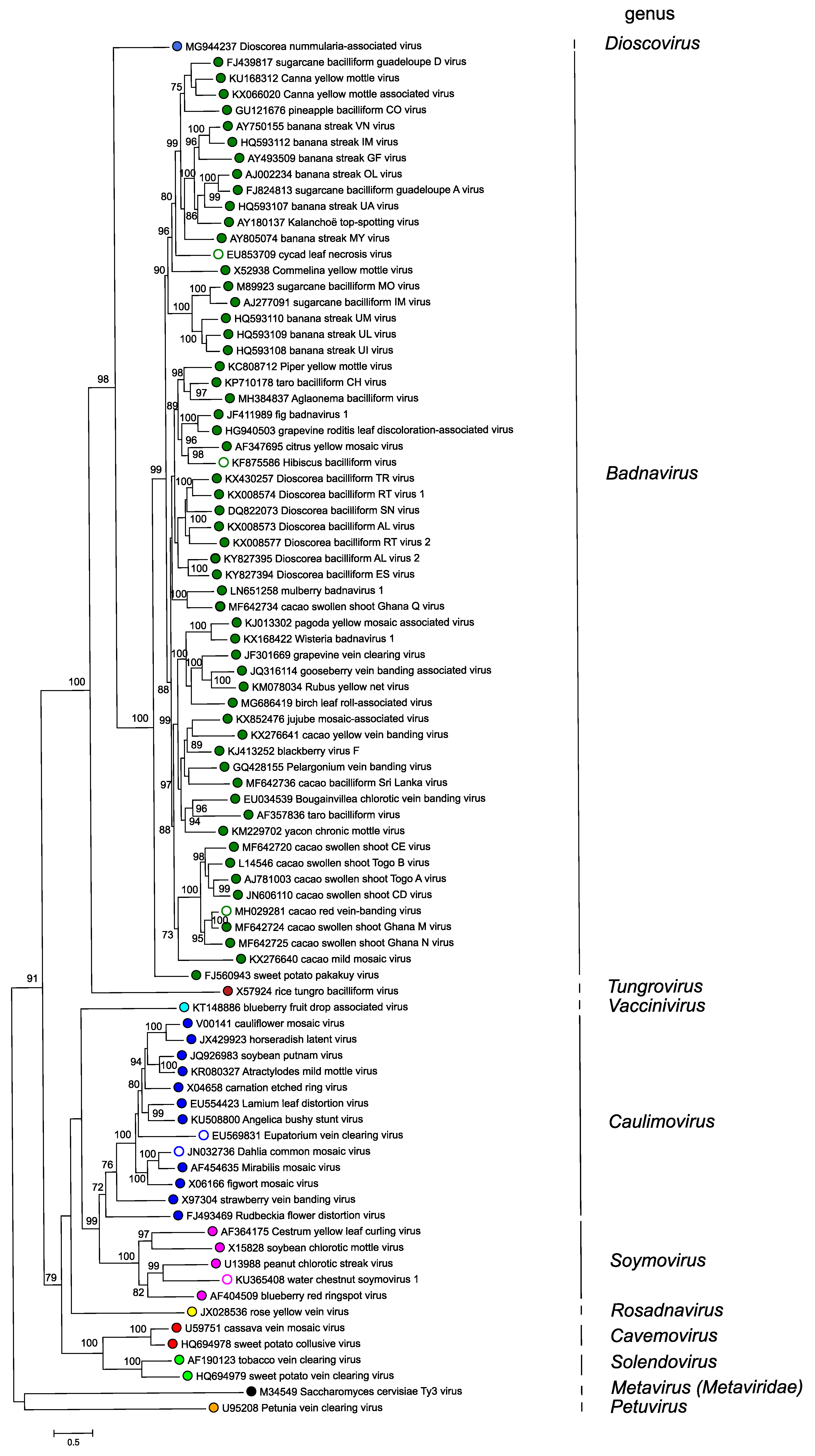
Phylogenetic relationships within the family have not yet been fully resolved and some of the relationships that are deduced depend on the method of analysis. However, several systematic studies have concluded that the genus Petuvirus is sister to all other genera in the Caulimoviridae and that all genera in the family Caulimoviridae form a monophyletic group that is sister to the family Metaviridae (Figure 4. Caulimoviridae).
 |
|
Figure 4. Caulimoviridae. Phylogenetic tree showing inferred evolutionary relationships of viruses in different genera in the family Caulimoviridae. Polymerase gene sequences of exemplar virus isolates of each of the species, homologous to nucleotide positions 3741–5654 of cauliflower mosaic virus (V00141) were aligned as conceptual amino acid sequences using PROMALS3D, guided by PDB protein structural files 3S8I_A, 4Z2Z_A, 4OL8_E and 4MH8_A. Corresponding DNA sequence alignments were then generated using TRANALIGN. A maximum likelihood estimate of phylogeny was done using IQTree v. 1.7 beta. Support values from an approximate likelihood ratio test with 10,000 replicates and ultrafast bootstrap with 1,000 replicates are shown at nodes of the branches. Saccharomyces cerevisiae Ty3 virus (genus Metavirus, family Metaviridae), was used as an outgroup. Colored dots indicate genera, with open circles indicating unclassified viruses in a genus. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Members of the family Caulimoviridae have the conserved gag-pol replication core of all reverse-transcribing viruses in the order Ortervirales (Krupovic et al., 2018). The domain architectures of the capsid protein, aspartic protease and reverse transcriptase proteins are similar to those of retroviruses (Krupovic and Koonin 2017). An additional unifying characteristic of the Ortervirales is a cytosolic tRNA priming mechanism for reverse transcription.
Members of the Caulimoviridae are frequently referred to as being ‘plant-infecting pararetroviruses’ (Hohn and Richert-Pöggeler 2006). The term ‘pararetrovirus’ was coined to describe reverse-transcribing viruses that do not integrate in the host genome as part of the replication cycle and which encapsidate dsDNA instead of ssRNA (Temin 1989). However, the two ‘pararetrovirus’ families, Hepadnaviridae and Caulimoviridae, are distantly related in phylogenetic studies using the conserved polymerase gene sequences and also differ in several other important ways: hepadnaviruses use a protein priming mechanism for reverse transcription and lack an aspartic protease enzyme.